7. Figure S5
[1]:
import os
import anndata
from pysankey import sankey # install via: pip install pysankeybeta
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import matplotlib as mpl
mpl.rcParams['pdf.fonttype'] = 42
mpl.rcParams['ps.fonttype'] = 42
mpl.rcParams['font.size'] = 12
mpl.rcParams['font.family'] = 'sans-serif'
mpl.rcParams['font.sans-serif'] = 'Helvetica'
dpi = 300
def plot_overlap_heatmap(df, col_y, col_x, y_order=None, x_order=None, cmap="Blues", figsize=(6, 6),
invert_y=True, invert_x=True):
# contingency table
ct = pd.crosstab(df[col_y], df[col_x])
# apply explicit orders or frequency sort
if y_order is not None:
y_idx = [i for i in y_order if i in ct.index]
else:
y_idx = ct.sum(axis=1).sort_values(ascending=False).index.tolist()
if invert_y:
y_idx = y_idx[::-1]
ct = ct.reindex(index=y_idx)
if x_order is not None:
x_idx = [i for i in x_order if i in ct.columns]
else:
x_idx = ct.sum(axis=0).sort_values(ascending=False).index.tolist()
if invert_x:
x_idx = x_idx[::-1]
ct = ct.reindex(columns=x_idx)
# row-normalize to percentages
data = ct.div(ct.sum(axis=1), axis=0).fillna(0) * 100
fig, ax = plt.subplots(figsize=figsize)
im = ax.imshow(data.values, aspect='equal', cmap=cmap, vmin=0, vmax=100)
ax.set_xticks(np.arange(data.shape[1]))
ax.set_yticks(np.arange(data.shape[0]))
ax.set_xticklabels(data.columns, rotation=90)
ax.set_yticklabels(data.index,)
ax.set_xlabel(col_x)
ax.set_ylabel(col_y)
cbar = fig.colorbar(im, ax=ax, fraction=0.046, pad=0.04)
cbar.set_label('% of source celltype')
fig.tight_layout()
return fig, ax
[2]:
path_full_h5ad = "../figure_data/input_data_Xenium/xenium_ovarian_cancer_full.h5ad"
output_folder = "../manuscript_figures/Figure_S5/"
os.makedirs(output_folder, exist_ok = True)
[3]:
adata = anndata.read_h5ad(path_full_h5ad)
[4]:
no_tumor_adata = adata[~adata.obs["is_tumor_cell_type"]]
no_tumor_adata_test = no_tumor_adata[no_tumor_adata.obs.is_in_vitmae_test_set]
no_tumor_adata_test = no_tumor_adata_test[~no_tumor_adata_test.obs["10X_cell_type"].isna(), :]
columns = ["10X_cell_type", 'SCimilarity_transcriptome_cell_type', 'SCimilarity_image_cell_type']
df = no_tumor_adata_test.obs.get(columns)
[5]:
cell_order_10x = [
'T and NK Cells',
'Stromal Associated Fibroblasts',
'Stromal Associated Endothelial Cells',
'Fallopian Tube Epithelium',
'Ciliated Epithelial Cells',
'Unassigned',
'Macrophages',
'Pericytes',
'Granulosa Cells',
'Smooth Muscle Cells',
]
cell_order_transcriptome = [
'leukocyte',
'stromal cell of ovary',
'fibroblast',
'endothelial cell',
'epithelial cell',
'mast cell',
'macrophage',
'dendritic cell',
'myofibroblast cell',
'monocyte',
'smooth muscle cell',
]
cell_order_image = [x for x in cell_order_transcriptome if x not in ['stromal cell of ovary', 'mast cell']]
cell_order_10x.reverse()
cell_order_transcriptome.reverse()
cell_order_image.reverse()
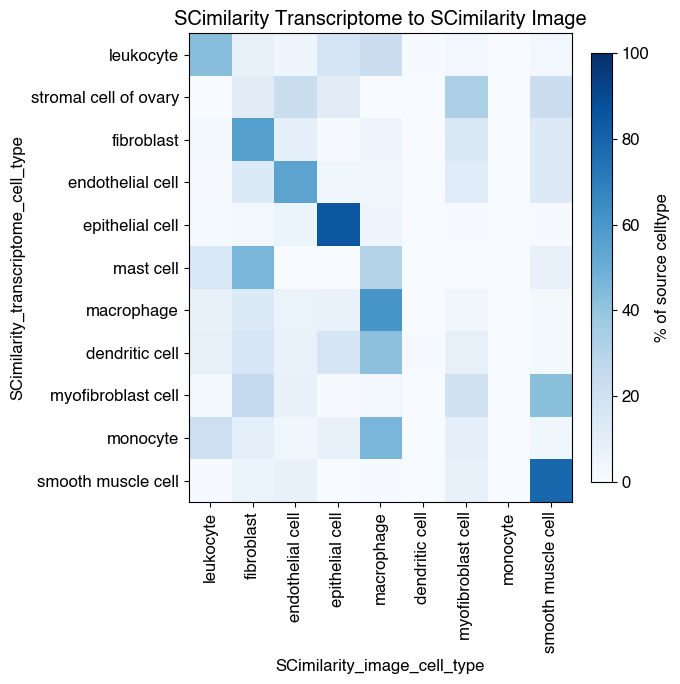
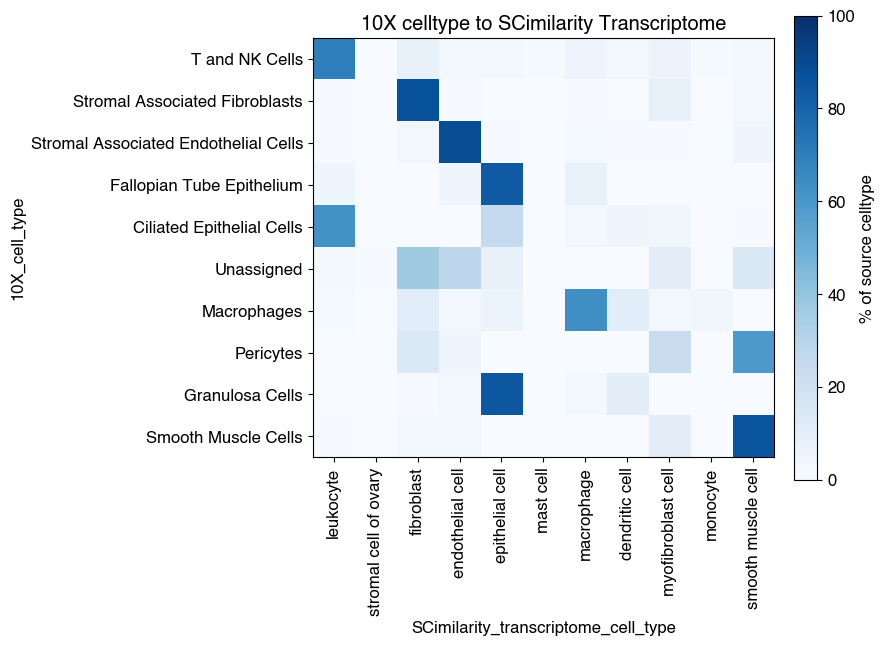
7.1. Fig S5a: quantify degree of overlap 10X cell type labels and SCimilarity transcriptome cell type labels
[6]:
fig, ax = plot_overlap_heatmap(
df,
col_y='10X_cell_type',
col_x='SCimilarity_transcriptome_cell_type',
y_order=cell_order_10x,
x_order=cell_order_transcriptome,
figsize=(9, 7)
)
ax.set_title('10X celltype to SCimilarity Transcriptome')
fig.tight_layout()
fig.savefig(f"{output_folder}/FigS5a_heatmap_celltype_transitions_10X_to_SCimilarity_transcriptome.pdf", dpi = 300)
1 extra bytes in post.stringData array
'created' timestamp seems very low; regarding as unix timestamp
7.2. Fig S5b: quantify degree of overlap SCimilarity transcriptome cell type labels and SCimilarity image cell type labels
[7]:
fig, ax = plot_overlap_heatmap(
df,
col_y='SCimilarity_transcriptome_cell_type',
col_x='SCimilarity_image_cell_type',
y_order = cell_order_transcriptome,
x_order = cell_order_image,
figsize=(7, 7)
)
ax.set_title('SCimilarity Transcriptome to SCimilarity Image')
fig.tight_layout()
fig.savefig(f"{output_folder}/FigS5b_heatmap_celltype_transitions_SCimilarity_transcriptome_to_SCimilarity_image.pdf", dpi = 300)
1 extra bytes in post.stringData array
'created' timestamp seems very low; regarding as unix timestamp