SPARCSpy - image-based single cell analysis at scale in python
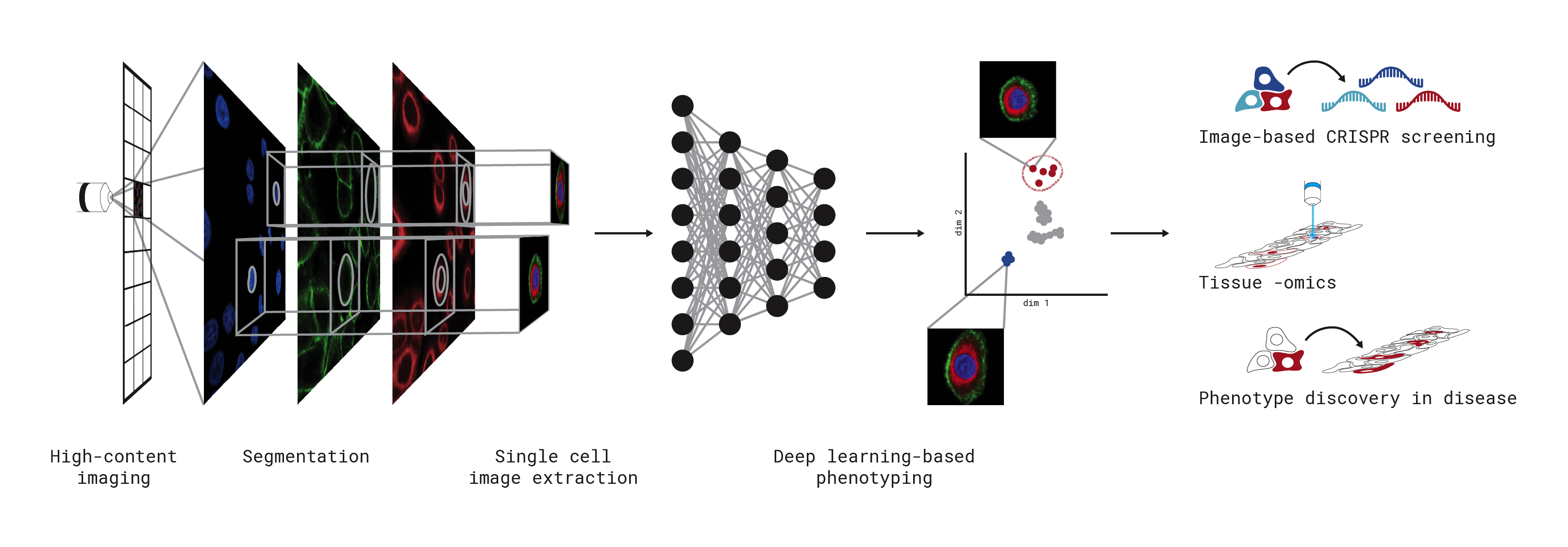
SPARCSpy is a scalable toolkit to analyse single-cell image datasets. This Python implementation efficiently segments individual cells, generates single-cell datasets and provides tools for the efficient deep learning classification of their phenotypes for downstream applications.
Installation
Check out the installation instructions here. You can validate your installation by running one of the example notebooks here.
Getting Started
You can check out our quickstart guide to get started with SPARCSpy. For more detailed information on the package, we have written an in depth computational workflow guide. In the github repository you can also find some tutorial notebooks as well as small example datasets to get started with. If you encounter issues feel free to open up a git issue.
Citing our Work
SPARCSpy was developed by Sophia Mädler, Georg Wallmann and Niklas Schmacke in the labs of Matthias Mann and Veit Hornung in 2023. SPARCSpy is actively developed with support from the labs of Matthias Mann, Veit Hornung and Fabian Theis.
If you use our code please cite the following manuscript:
SPARCS, a platform for genome-scale CRISPR screening for spatial cellular phenotypes Niklas Arndt Schmacke, Sophia Clara Maedler, Georg Wallmann, Andreas Metousis, Marleen Berouti, Hartmann Harz, Heinrich Leonhardt, Matthias Mann, Veit Hornung bioRxiv 2023.06.01.542416; doi: https://doi.org/10.1101/2023.06.01.542416
Contributing
We are excited for people to adapt and extend SPARCSpy to their needs. If you are interested in contributing to SPARCSpy, please reach out to the developers or open a pull request on our github repository.
Documentation
SPARCSpy Ecosystem