A walk through the SPARCSpy Ecosystem
This notebook will introduce you to the SPARCSpy ecosystem and give you a complete working example of how to use SPARCSpy. It will walk you through the following steps of the SPARCSpy workflow: 1. segmentation: Generates masks for the segmentation of input images into individual cells 2. extraction: The segmentation masks are applied to extract single-cell images for all cells in the input images. Images of individual cells are rescaled to [0, 1] per channel. 3. classification: The
image-based phenotype of each individual cell in the extracted single-cell dataset is classified using the specified classification method. Multiple classification runs can be performed on the same dataset using different classification methods. Here we utilize the pretrained binary classifier from the original SPARCS manuscript that identifies individual cells defective in a biological process called “autophagy”. 4. selection: Cutting
instructions for the isolation of selected individual cells by laser microdissection are generated. The cutting shapes are written to an .xml file which can be loaded on a leica LMD microscope for automated cell excision.
The data used in this notebook was previously stitched using the stitching workflow in SPARCStools. Please see the notebook here.
Import Required Libraries
First we need to import all of the python functions we require to run the pipeline.
[2]:
import os
import numpy as np
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import pandas as pd
from sparcscore.pipeline.project import Project
from sparcscore.pipeline.workflows import WGASegmentation
from sparcscore.pipeline.extraction import HDF5CellExtraction
from sparcscore.pipeline.classification import MLClusterClassifier
from sparcscore.pipeline.selection import LMDSelection
Generate the Project Structure
Executing this code will generate a new SPARCSpy project in the designated location. Each SPARCSpy project has the following general structure:
.
├── classification
│ └── classifier_name
│ └── processing.log
├── config.yml
├── extraction
├── segmentation
└── processing.log
[3]:
project_location = f"../../../example_data/example_1/project"
project = Project(os.path.abspath(project_location),
config_path= "config_test.yml",
overwrite=False,
debug=True,
segmentation_f=WGASegmentation,
extraction_f=HDF5CellExtraction,
classification_f=MLClusterClassifier,
selection_f=LMDSelection
)
modifying config
[30/01/2024 11:59:34] Loading config from /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/config.yml
[30/01/2024 11:59:34] current run: 0
/Users/sophia/Documents/GitHub/SPARCSpy/src/sparcscore/pipeline/project.py:109: UserWarning: Theres already a directory in the location path
warnings.warn("Theres already a directory in the location path")
Load Imaging Data
Once we have generated our project structure we next need to load our imaging data. There are several different ways to do this.
you can load the images directly from file by specifying a list of filepaths
you can load the images from numpy arrays that are already loaded into memory
Here it is important that you load the channels in the following order: Nucleus, Cellmembrane, others
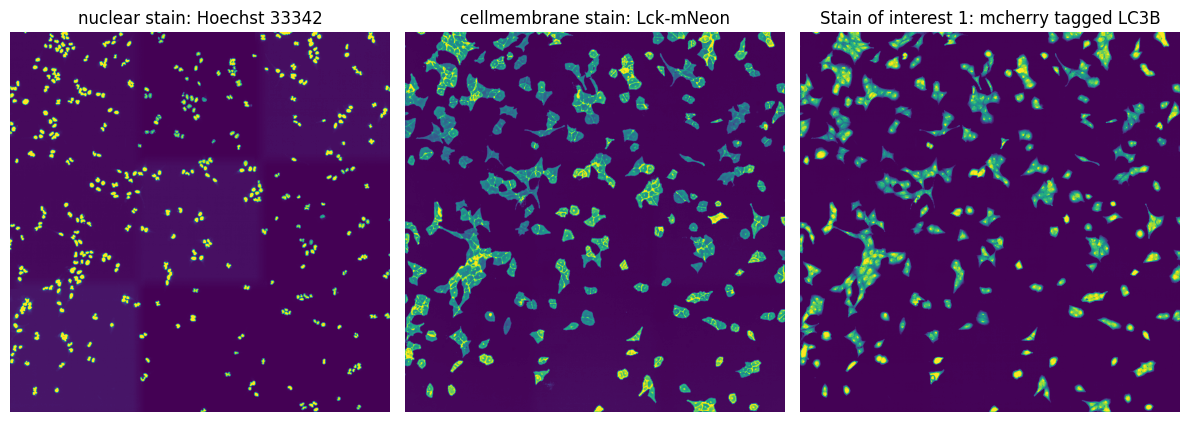
In this particular example we are utilizing images from U2OS cells which are stained with Hoechst 33342 to visualize nuclei and express Lck-mNeon to visualize the cellular membrane and LC3B-mcherry, a fluroescently tagged protein relevant for visualizing the biological process of autophagy. The cells have not been stimulated to induce autophagy and should be autophagy defective.
Method 1: loading images from file
[4]:
images = ["../../../example_data/example_1/stitched_images/stitching_test_DAPI.tif",
"../../../example_data/example_1/stitched_images/stitching_test_Alexa488.tif",
"../../../example_data/example_1/stitched_images/stitching_test_mCherry.tif"]
project.load_input_from_file(images)
(3, 3039, 3037)
Method 2: loading images from numpy array
To simulate the case where the images you want to load are already loaded as a numpy array, we first convert the images to a numpy array and then pass this array to the project instead of only providing the image paths.
[5]:
from tifffile import imread
images = ["../../../example_data/example_1/stitched_images/stitching_test_DAPI.tif",
"../../../example_data/example_1/stitched_images/stitching_test_Alexa488.tif",
"../../../example_data/example_1/stitched_images/stitching_test_mCherry.tif"]
image_arrays = np.array([imread(path) for path in images])
project.load_input_from_array(image_arrays)
Looking at the loaded images
The loaded images are accesible via the input_image parameter of the project and are saved as numpy arrays with the following structure (c, x, y)
[6]:
project.input_image
[6]:
array([[[0.000e+00, 0.000e+00, 0.000e+00, ..., 2.393e+03, 2.404e+03,
0.000e+00],
[0.000e+00, 1.494e+03, 1.253e+03, ..., 2.349e+03, 2.209e+03,
0.000e+00],
[0.000e+00, 1.343e+03, 1.449e+03, ..., 2.593e+03, 2.386e+03,
0.000e+00],
...,
[0.000e+00, 4.092e+03, 4.057e+03, ..., 2.300e+02, 2.170e+02,
0.000e+00],
[0.000e+00, 4.620e+03, 4.303e+03, ..., 2.540e+02, 2.190e+02,
0.000e+00],
[0.000e+00, 0.000e+00, 0.000e+00, ..., 0.000e+00, 0.000e+00,
0.000e+00]],
[[0.000e+00, 0.000e+00, 0.000e+00, ..., 8.720e+02, 6.590e+02,
0.000e+00],
[0.000e+00, 3.100e+02, 4.560e+02, ..., 5.950e+02, 2.730e+02,
0.000e+00],
[0.000e+00, 4.710e+02, 3.120e+02, ..., 6.360e+02, 4.410e+02,
0.000e+00],
...,
[0.000e+00, 9.950e+02, 9.030e+02, ..., 1.089e+03, 1.293e+03,
0.000e+00],
[0.000e+00, 9.890e+02, 8.530e+02, ..., 1.369e+03, 1.586e+03,
0.000e+00],
[0.000e+00, 0.000e+00, 0.000e+00, ..., 0.000e+00, 0.000e+00,
0.000e+00]],
[[0.000e+00, 0.000e+00, 0.000e+00, ..., 7.830e+02, 1.122e+03,
0.000e+00],
[0.000e+00, 1.000e+00, 2.000e+00, ..., 8.820e+02, 9.590e+02,
0.000e+00],
[0.000e+00, 4.400e+01, 6.500e+01, ..., 8.560e+02, 8.060e+02,
0.000e+00],
...,
[0.000e+00, 1.290e+02, 9.200e+01, ..., 6.340e+02, 3.120e+02,
0.000e+00],
[0.000e+00, 4.440e+02, 2.320e+02, ..., 4.690e+02, 2.650e+02,
0.000e+00],
[0.000e+00, 0.000e+00, 0.000e+00, ..., 0.000e+00, 0.000e+00,
0.000e+00]]])
[9]:
# visualize input images as example
# it is not recommended to execute this block with large input images as it will compute for some time
fig, axs = plt.subplots(1, 3, figsize = (12, 6));
axs[0].imshow(project.input_image[0]);
axs[0].axis("off");
axs[0].set_title("nuclear stain: Hoechst 33342")
axs[1].imshow(project.input_image[1]);
axs[1].axis("off");
axs[1].set_title("cellmembrane stain: Lck-mNeon")
axs[2].imshow(project.input_image[2]);
axs[2].axis("off");
axs[2].set_title("Stain of interest 1: mcherry tagged LC3B")
fig.tight_layout()
Generating a Segmentation
SPARCSpy has different segmentation workflows between which you can choose. If none of them fit your needs you can also easily write your own.
Here we will demonstrate the CPU based classical segmentation method that was also utilized in the manuscript.
We define the segmentation method to be used when we initialize the project. In this case we used the WGASegmentation method. The ShardedWGASegmentation works very similarily but allows you to process several image chunks in parallel for more efficient computation on large input images.
By specifying debug = True we can see intermediate output results from the segmentation.
The WGASegmentation method relies on parameters specified in the config.yml we loaded when initializing our project.
WGASegmentation:
input_channels: 3
chunk_size: 50 # chunk size for chunked HDF5 storage
lower_quantile_normalization: 0.001
upper_quantile_normalization: 0.999
median_filter_size: 4 # Size in pixels
nucleus_segmentation:
lower_quantile_normalization: 0.01
upper_quantile_normalization: 0.99
median_block: 41
median_step: 4
threshold: 0.2
min_distance: 8
peak_footprint: 7
speckle_kernel: 9
dilation: 0 # final dilation of pixel mask
min_size: 200 # minimum nucleus area in pixel
max_size: 5000 # maximum nucleus area in pixel
contact_filter: 0.5 # minimum nucleus contact with background
wga_segmentation:
threshold: 0.05
lower_quantile_normalization: 0.01
upper_quantile_normalization: 0.99
erosion: 2
dilation: 7
min_clip: 0
max_clip: 0.2
min_size: 200
max_size: 30000
chunk_size: 50
By passing the parameter debug = True we tell SPARCSpy to also generate intermediate outputs which we can look at to see the different segmentation steps. It is only recommended to do this for debugging or visualization purposes as it will utilize more memory and be slower.
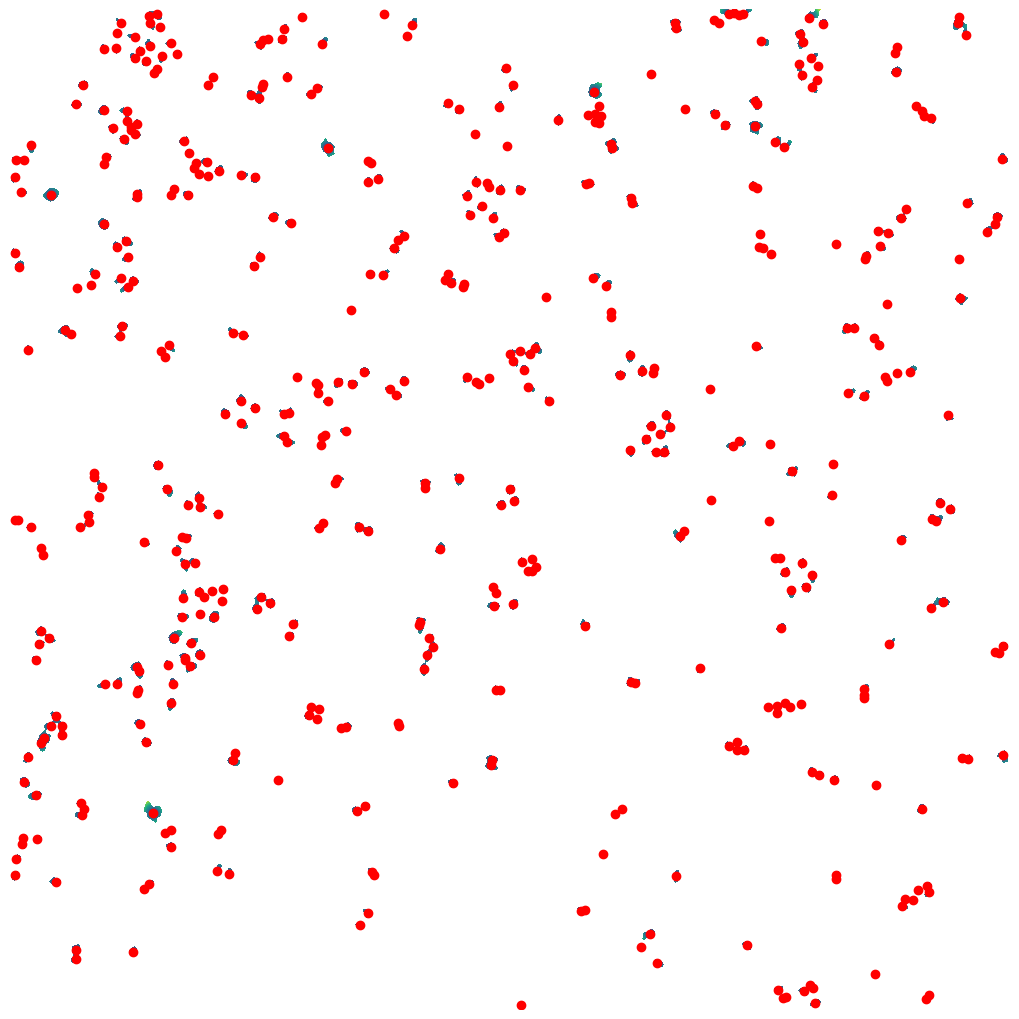
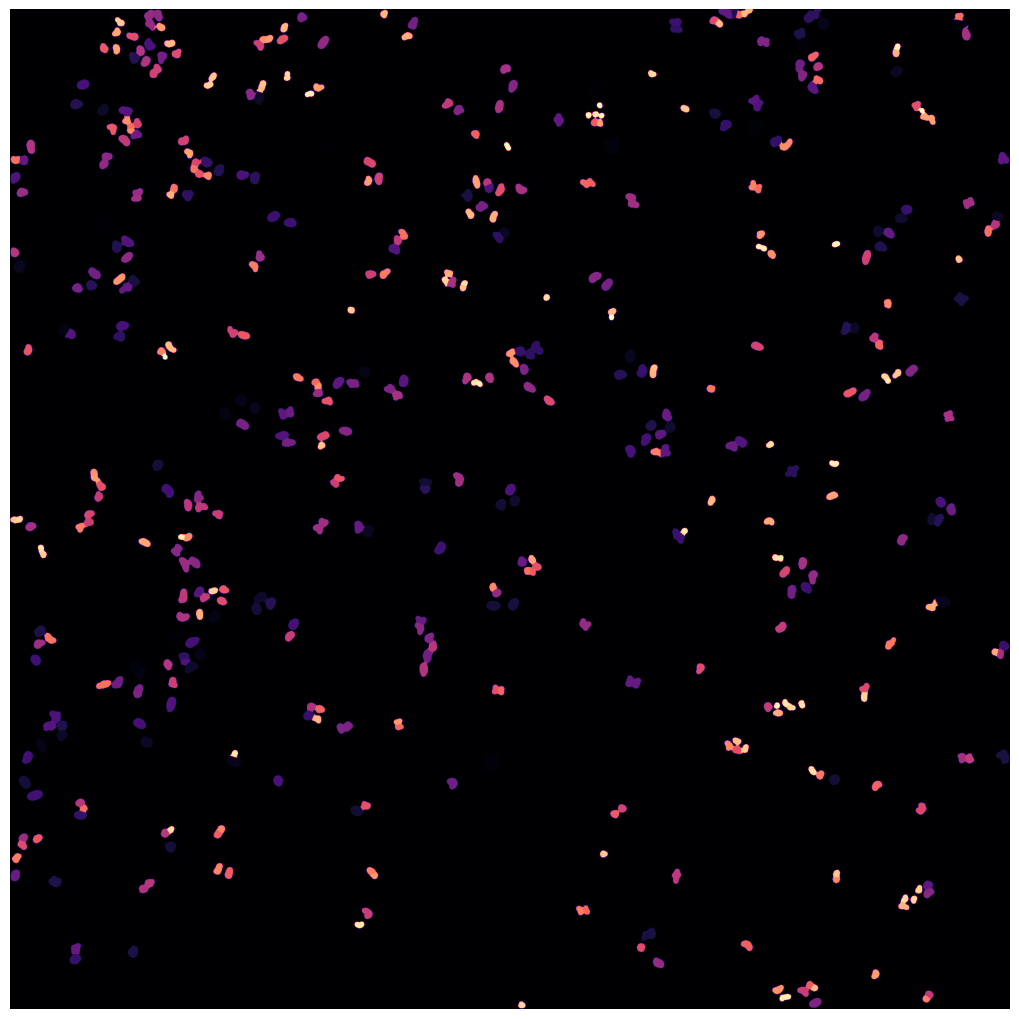
For the WGASegmentation method the intermediate outputs that are displayed are the following:
percentile normalized input images for each of the three channels (3 images)
median normalized input images (this slightl smooths the images for a better segmentation result) (3 images)
histogram showing the intensity distribution for nuclear and cytosolic channel (2 plots)
generated mask after applying nuclear thresholding
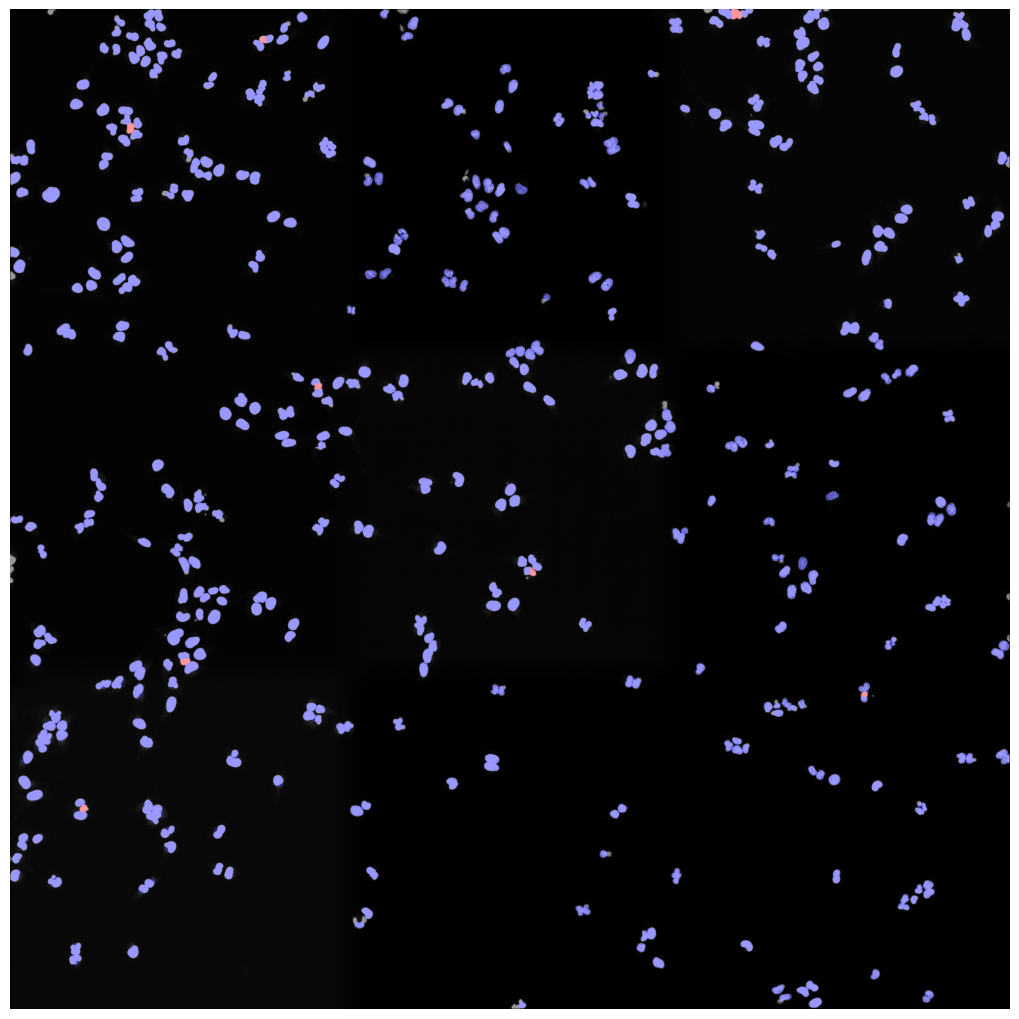
nuclear thresholding mask with calculated centers for each detected nucleus
fast marching map with nuclei centers indicated in red
segmented individual nuclei (2 different visualizations)
starting nucleus mask for filtering
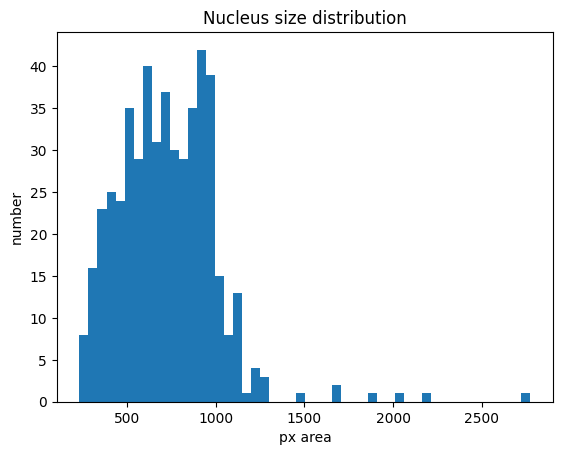
histrogram showing size distribution of segmented nuclei
segmentation mask with too small nuclei shown in red
segmentation mask with too large nuclei shown in red
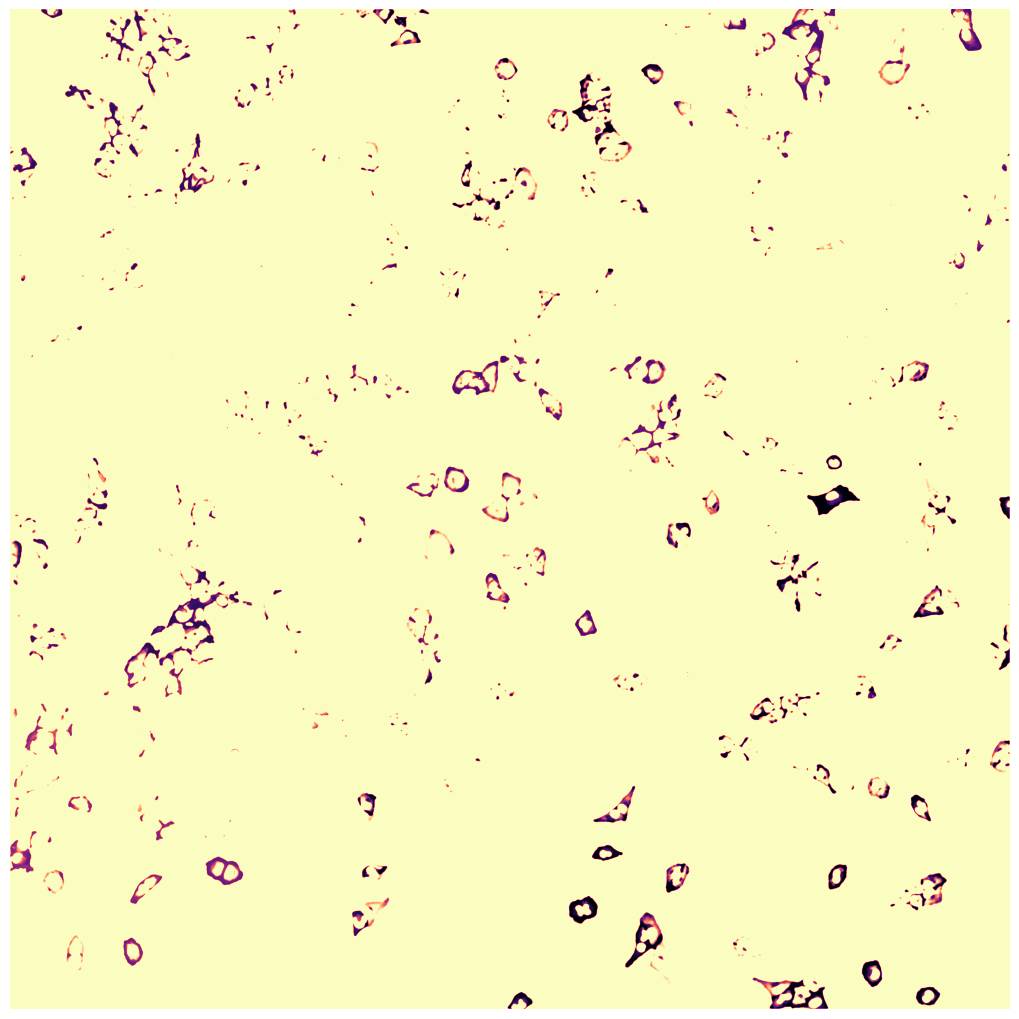
WGA thresholding mask
WGA potential map
WGA fast marching results
Watershed results with nuclear centers shown in red
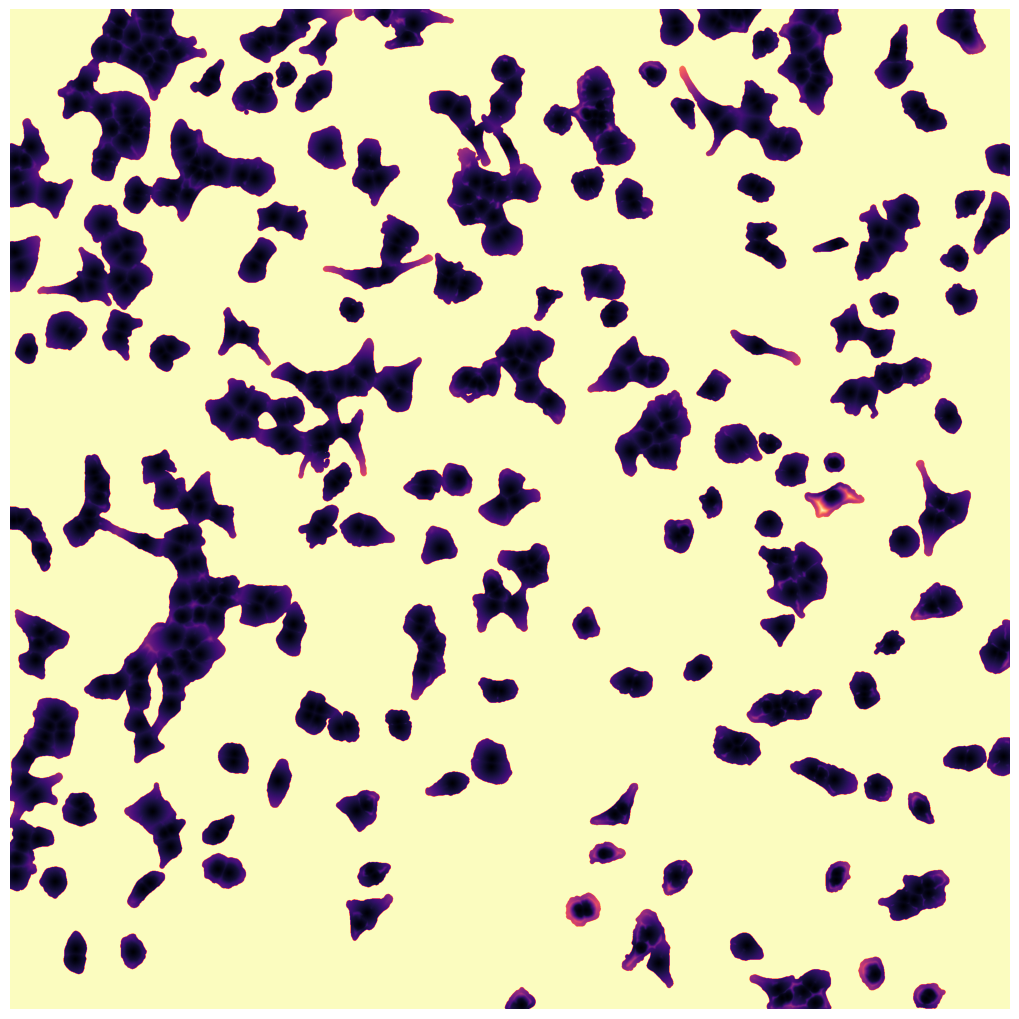
WGA mask
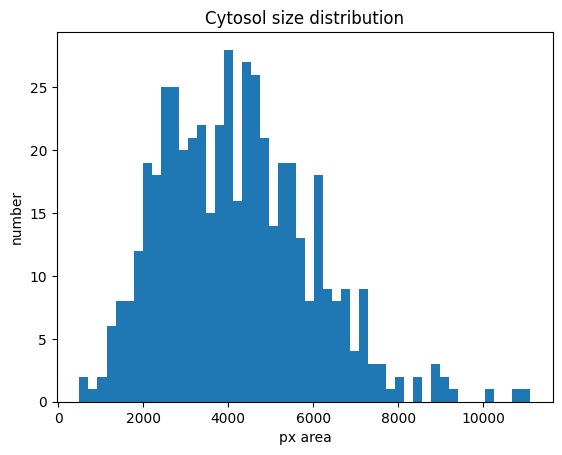
Cytosol size distribution
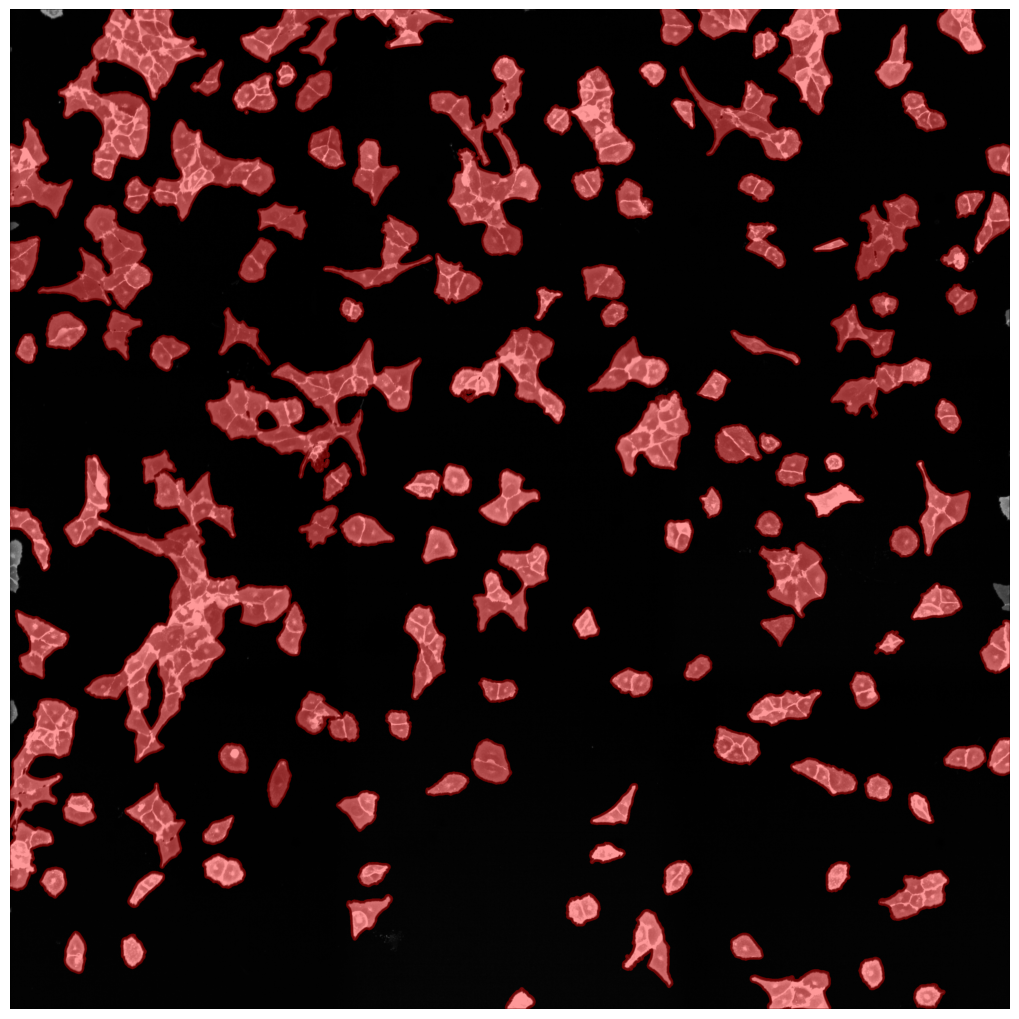
cytosol mask with too small cells shown in red
cytosol mask with too largec cells shown in red
[46]:
project.segment(debug = True)
[01/06/2023 14:49:35] No existing map 0 normalized found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/0_normalized_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 1 median found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/1_median_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 2 nucleus_segmentation found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/2_nucleus_segmentation_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 3 nucleus_mask found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/3_nucleus_mask_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 4 wga_mask found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/4_wga_mask_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 5 wga_potential found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/5_wga_potential_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 6 travel_time found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/6_travel_time_map.npy, new one will be created
[01/06/2023 14:49:35] No existing map 7 watershed found at path /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/7_watershed_map.npy, new one will be created
[01/06/2023 14:49:35] Segmentation started, starting from checkpoint 0
[01/06/2023 14:49:35] Started with normalized map
[01/06/2023 14:49:38] Normalized map created
[01/06/2023 14:49:38] Started with median map
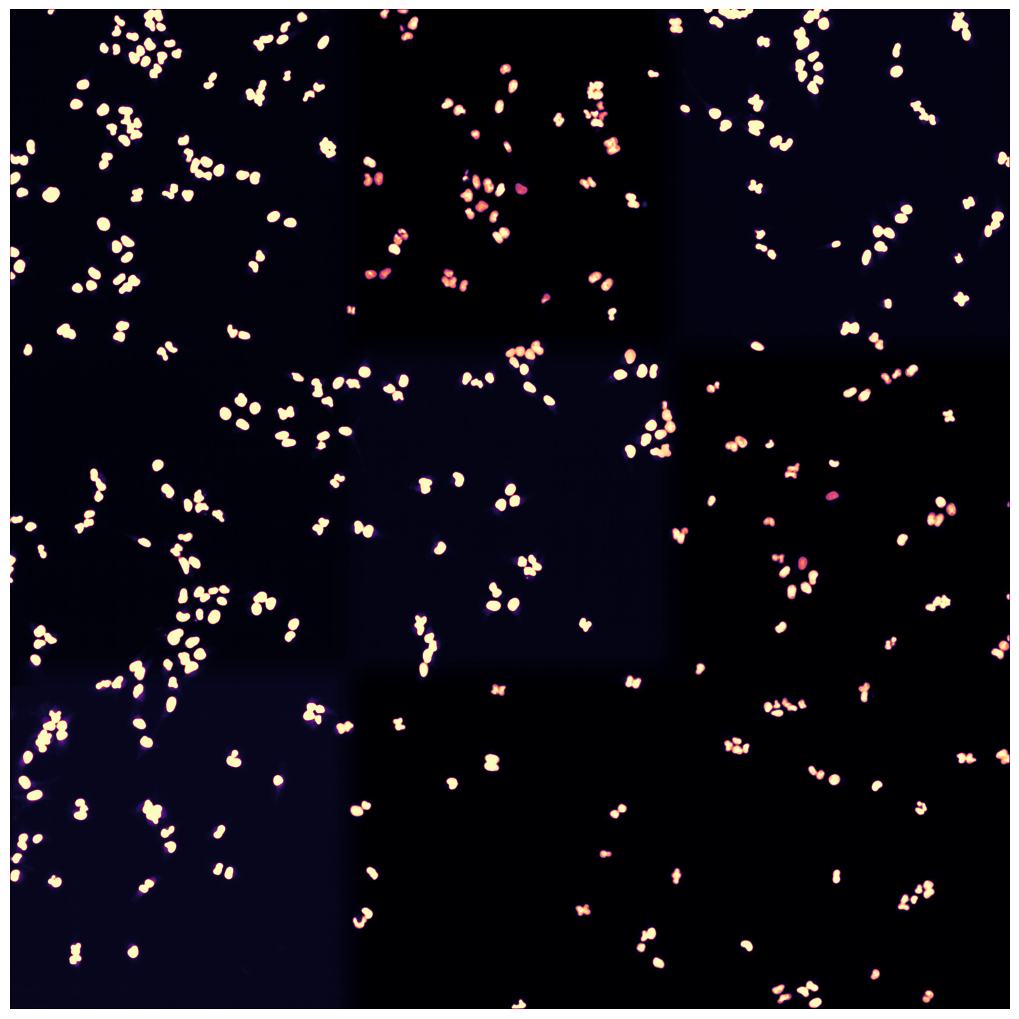
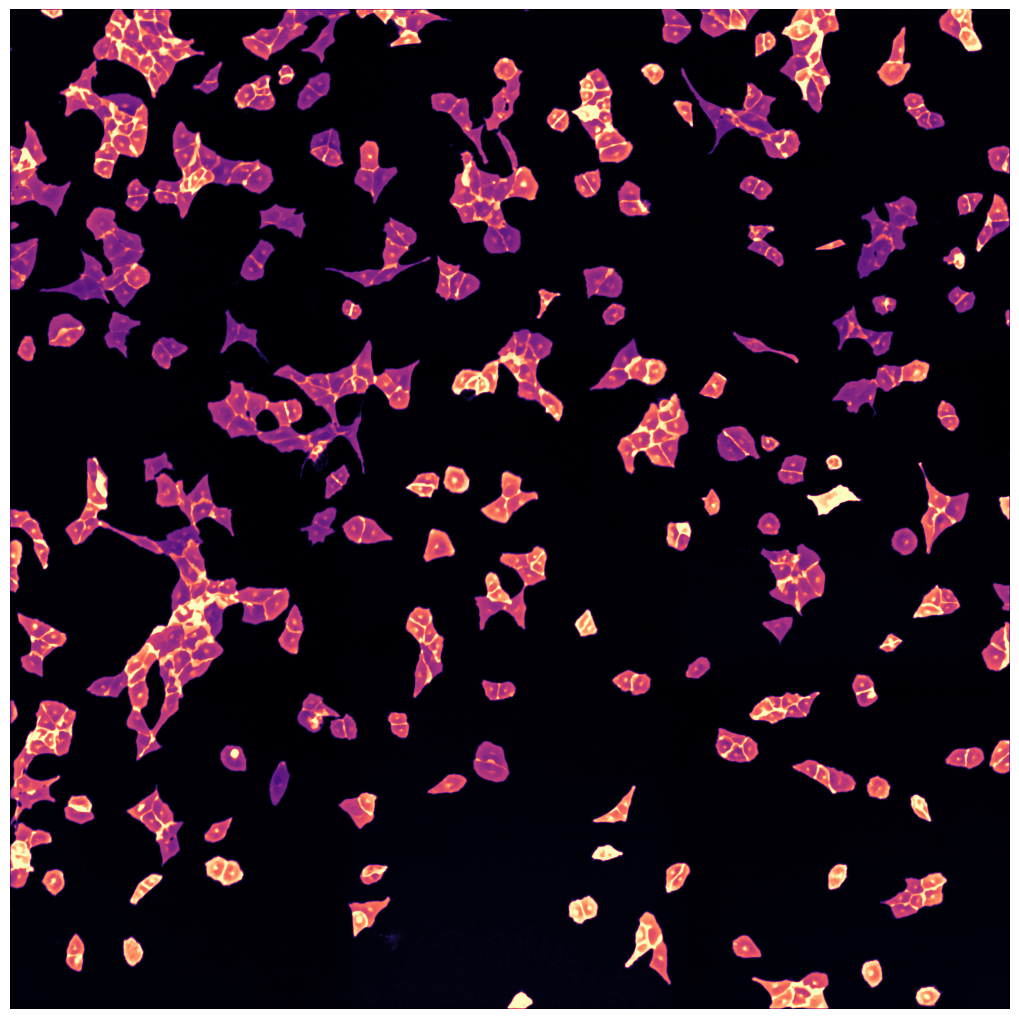
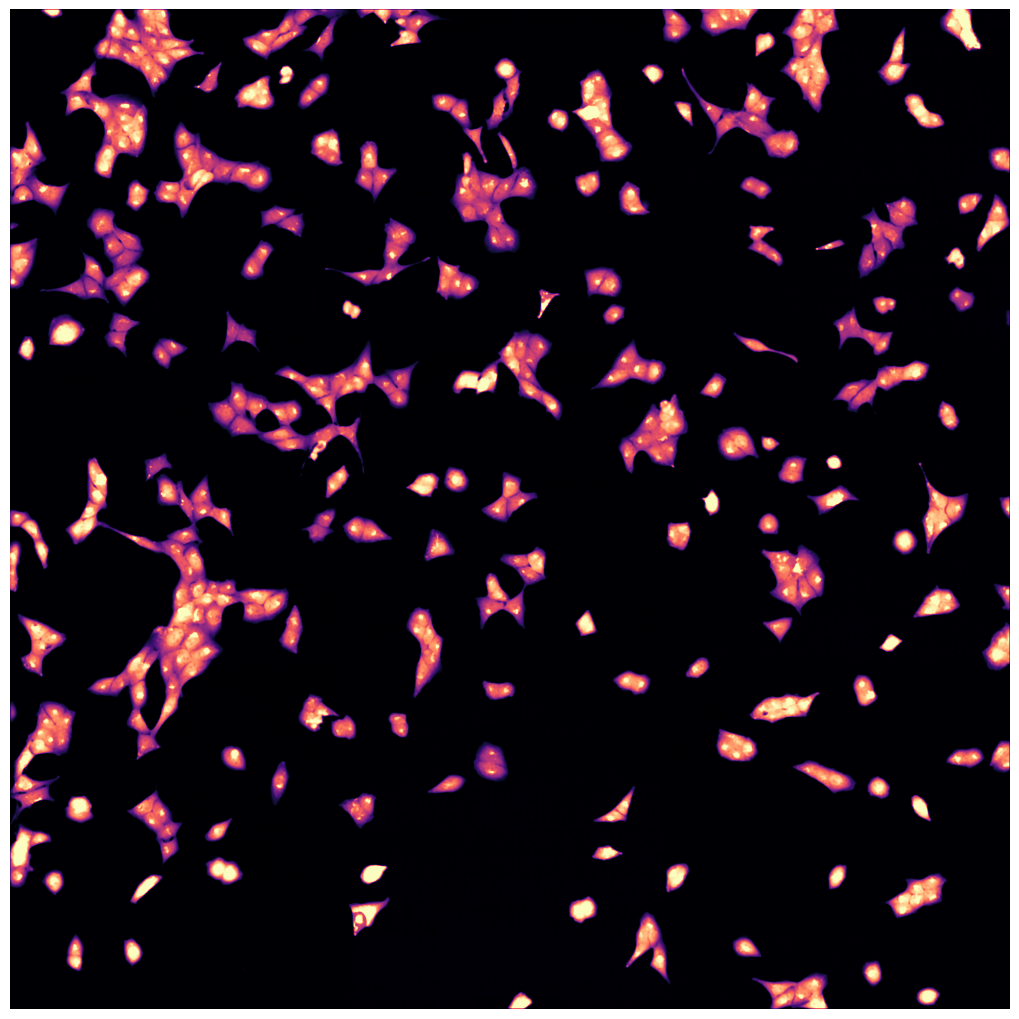
[01/06/2023 14:49:55] Median map created
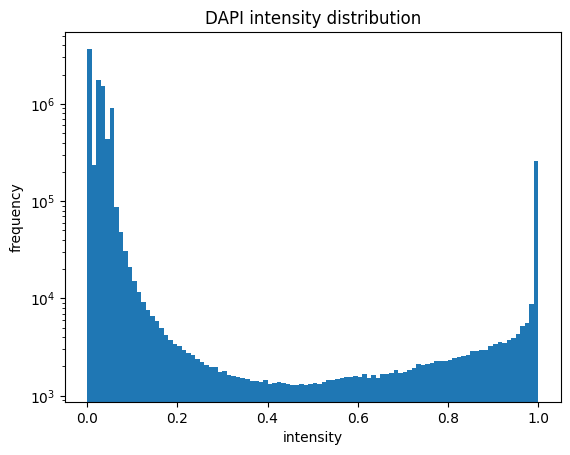
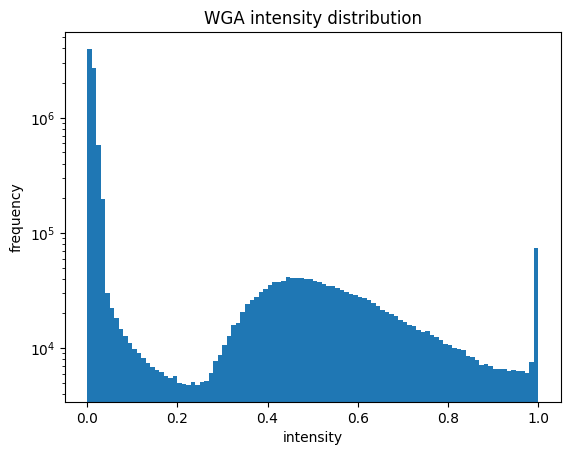
[01/06/2023 14:49:56] Started performing nucleus segmentation.
[01/06/2023 14:49:56] Generating thresholded nucleus map.

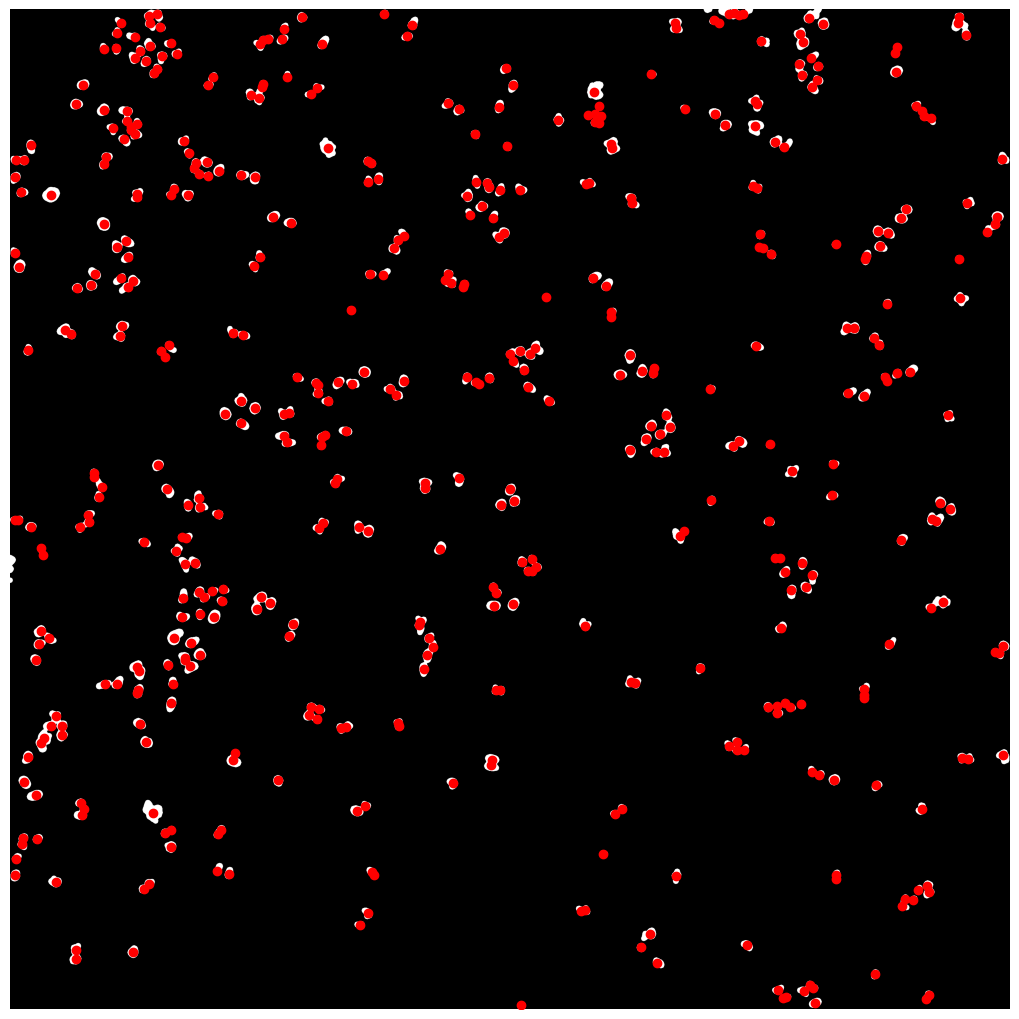

[01/06/2023 14:50:12] /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/2_nucleus_segmentation_map will be saved as tif
[01/06/2023 14:50:12] Thresholded nucleus map created.
[01/06/2023 14:50:12] Started with nucleus mask map
[01/06/2023 14:50:12] /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/3_nucleus_mask_map will be saved as tif

[01/06/2023 14:50:13] Nucleus mask map created with 494 elements
[01/06/2023 14:50:13] Filtered out due to contact limit: 10
[01/06/2023 14:50:14] Filtered out due to size limit: 0
[01/06/2023 14:50:14] Filtered out: 10

[01/06/2023 14:50:16] Started with WGA mask map

[01/06/2023 14:50:18] WGA mask map created
[01/06/2023 14:50:18] Started with WGA potential map
[01/06/2023 14:50:19] WGA mask potential created
[01/06/2023 14:50:19] Started with fast marching
/Users/sophia/Documents/GitHub/SPARCSpy/src/sparcscore/pipeline/workflows.py:191: RuntimeWarning: invalid value encountered in cast
px_center = np.round(center_nuclei).astype(int)
[01/06/2023 14:50:21] Fast marching finished
[01/06/2023 14:50:21] Started with watershed
/Users/sophia/Documents/GitHub/SPARCSpy/src/sparcscore/pipeline/workflows.py:213: RuntimeWarning: invalid value encountered in cast
px_center = np.round(center_nuclei).astype(int)
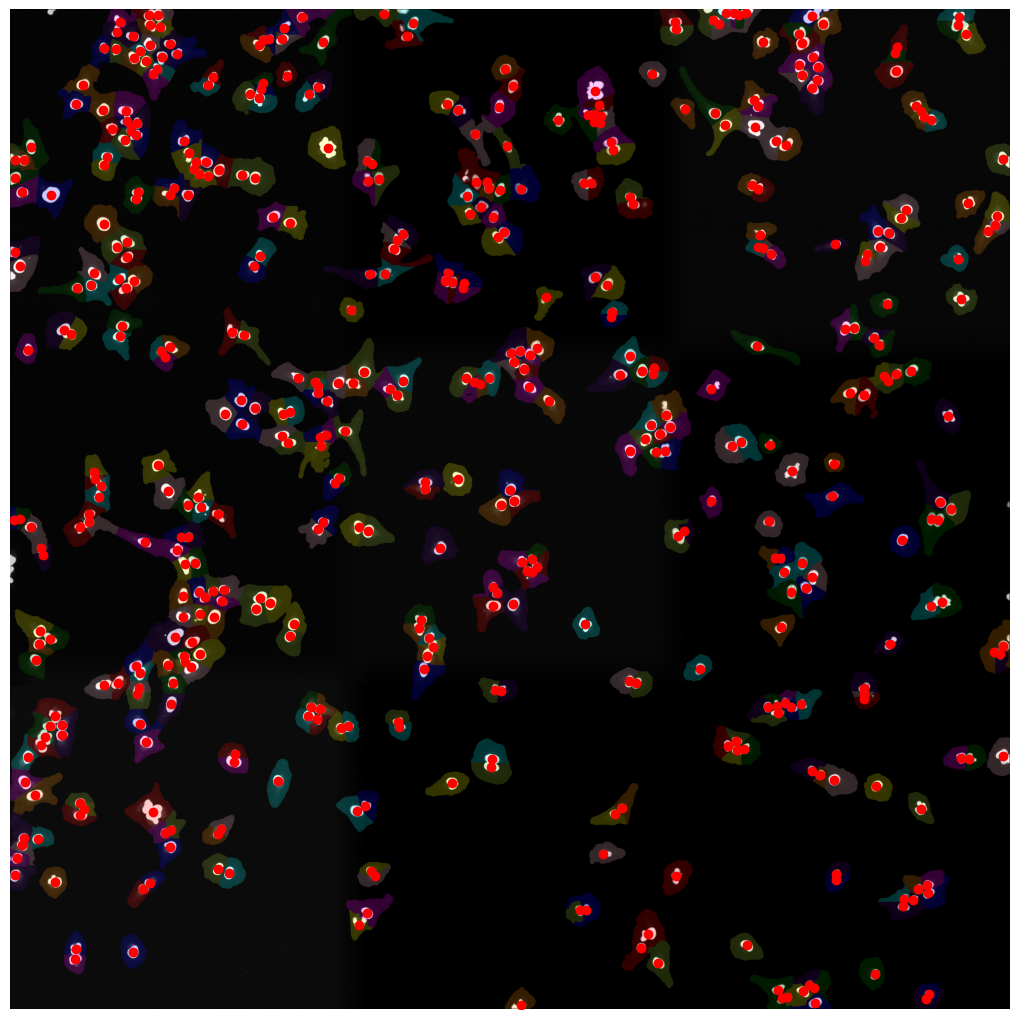
[01/06/2023 14:50:23] /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/7_watershed_map will be saved as tif
[01/06/2023 14:50:23] watershed finished
[01/06/2023 14:50:24] Cells filtered out due to cytosol size limit: 0
[01/06/2023 14:50:24] Filtered out: 10
[01/06/2023 14:50:24] Remaining: 485
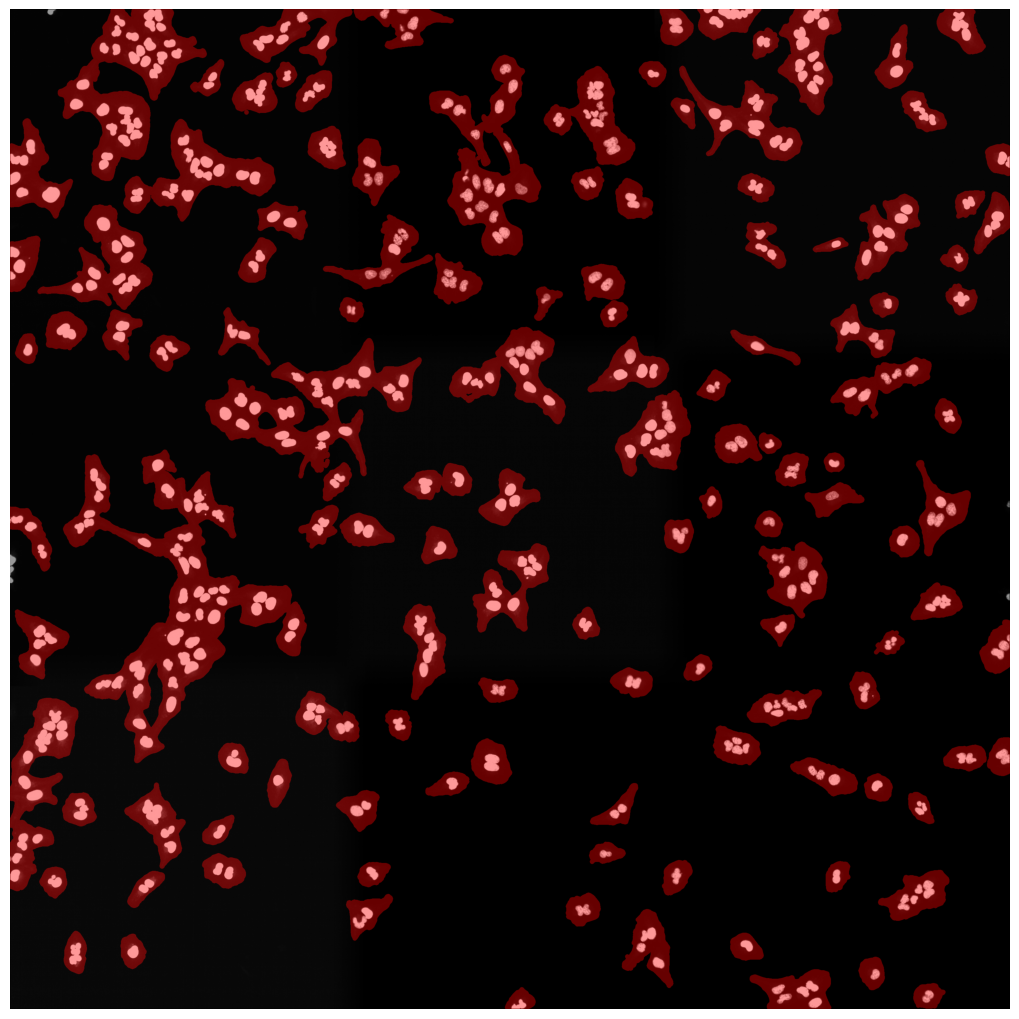
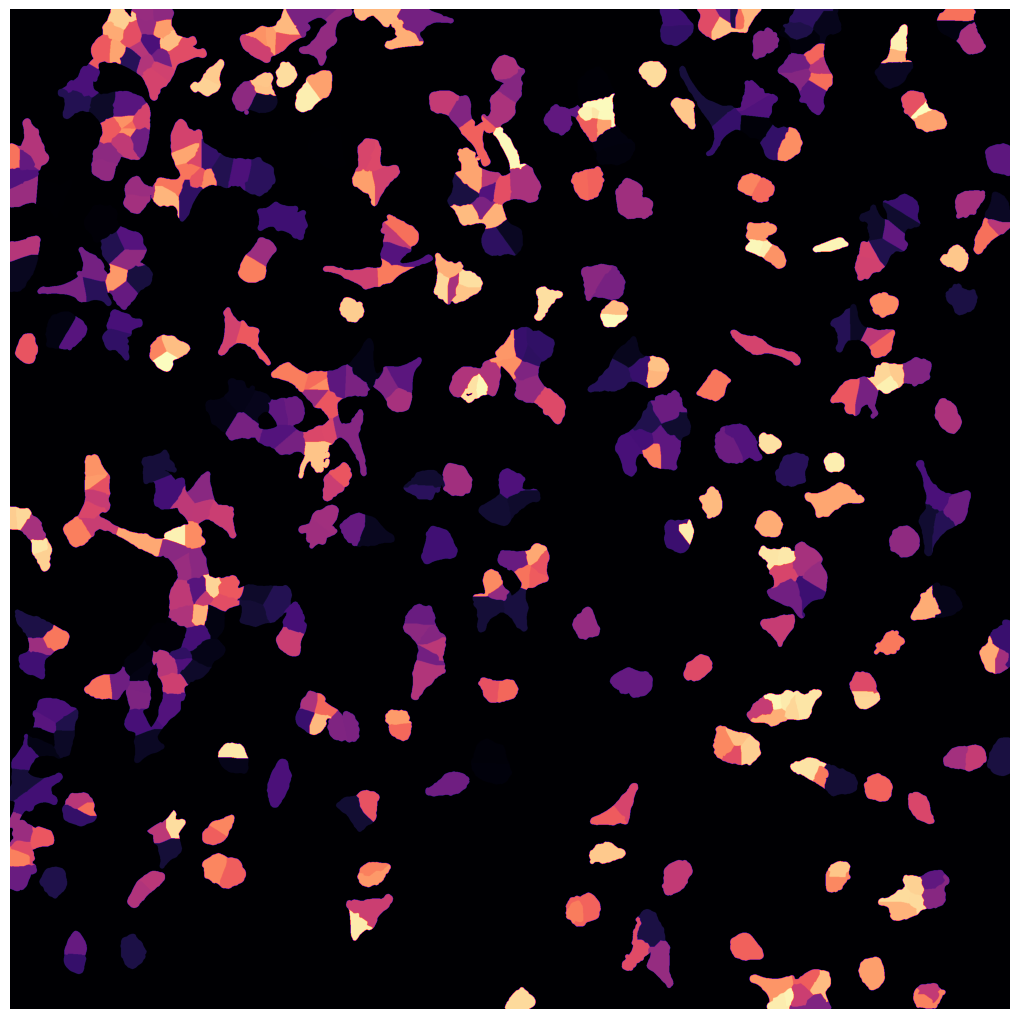
[01/06/2023 14:50:27] saving segmentation
[01/06/2023 14:50:27] === finished segmentation ===
Looking at Segmentation Results
The Segmentation Results are written to a hdf5 file called segmentation.h5 located in the segmentation directory of our SPARCSpy project.
The file contains two datasets: channels and labels. Channels contains the input images and labels the generated segmentation masks.
[47]:
import h5py
with h5py.File(f"{project_location}/segmentation/segmentation.h5") as hf:
print(hf.keys())
<KeysViewHDF5 ['channels', 'labels']>
If we look at the labels dataset we can see that it contains a numpy array containing two segmentation masks: the nuclear segmentation and the cytosol segmentation generated by our chosen segmentation method.
[48]:
with h5py.File(f"{project_location}/segmentation/segmentation.h5") as hf:
segmentation = hf.get("labels")
print(segmentation)
plt.figure()
plt.imshow(segmentation[0])
plt.axis("off")
plt.set_title("Nuclear Segmentation Mask")
plt.figure()
plt.imshow(segmentation[1])
plt.axis("off")
plt.set_title("Cytosol Segmentation Mask")
<HDF5 dataset "labels": shape (2, 3040, 3038), type "<i4">
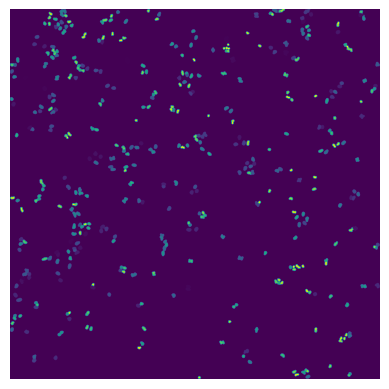
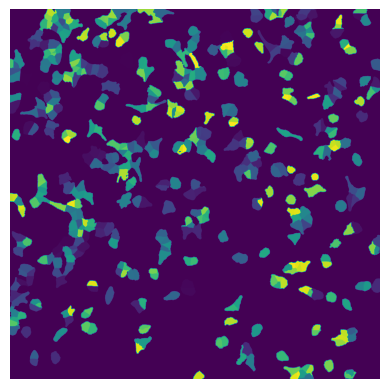
Each segmentation mask is an array where each pixel is assigned either to background (0) or to a specific cell (cellid: whole number).
If we zoom in on the corner of the segmentation mask of a nucleus the numpy array would look like this:
[49]:
with h5py.File(f"{project_location}/segmentation/segmentation.h5") as hf:
segmentation = hf.get("labels")
plt.imshow(segmentation[0, 80:90, 945:955])
plt.axis("off")
print(segmentation[0, 80:90, 945:955])
[[ 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 205]
[ 0 0 0 0 0 0 0 205 205 205]
[ 0 0 0 0 0 0 205 205 205 205]
[ 0 0 0 0 205 205 205 205 205 205]
[ 0 0 0 205 205 205 205 205 205 205]
[ 0 0 205 205 205 205 205 205 205 205]
[ 0 205 205 205 205 205 205 205 205 205]
[205 205 205 205 205 205 205 205 205 205]]

Extracting single-cell images
Once we have generated a segmentation mask, the next step is to extract single-cell images of segmented cells in the dataset.
Like during the segmentation there are several extraction methods to choose from. For regular SPARCSpy projects we need the HDF5CellExtraction method. This will extract single-cell images for all cells segmentated in the dataset and write them to a hdf5 file.
The parameters with which HDF5CellExtraction will run are again specified in the config.yml file.
HDF5CellExtraction:
compression: True
threads: 80 # threads used in multithreading
image_size: 128 # image size in pixel
cache: "."
hdf5_rdcc_nbytes: 5242880000 # 5gb 1024 * 1024 * 5000
hdf5_rdcc_w0: 1
hdf5_rdcc_nslots: 50000
[50]:
project.extract()
WARNING:root:Folder /var/folders/35/p4c58_4n3bb0bxnzgns1t7kh0000gn/T/./temp_mmap7udzn9ot with temp mmap arrays is being deleted.All existing temp mmapp arrays will be unusable!
WARNING:root:New temp folder location. Temp mmap arrays are written to /var/folders/35/p4c58_4n3bb0bxnzgns1t7kh0000gn/T/./temp_mmap24gm079o. Cleanup of this folder is OS dependant, and might need to be triggered manually!
[01/06/2023 14:50:28] Using channel label <HDF5 dataset "channels": shape (3, 3040, 3038), type "<f8">
[01/06/2023 14:50:28] Using segmentation label <HDF5 dataset "labels": shape (2, 3040, 3038), type "<i4">
[01/06/2023 14:50:28] Created new extraction cache ./1baf6da4-ccb6-46ec-bd51-5118a414bfa9
[01/06/2023 14:50:28] Started extraction
[01/06/2023 14:50:28] Loading segmentation data from {input_segmentation_path}
[01/06/2023 14:50:28] Using channel label <HDF5 dataset "channels": shape (3, 3040, 3038), type "<f8">
[01/06/2023 14:50:28] Using segmentation label <HDF5 dataset "labels": shape (2, 3040, 3038), type "<i4">
[01/06/2023 14:50:28] Finished loading channel data (3, 3040, 3038)
[01/06/2023 14:50:28] Finished loading label data (2, 3040, 3038)
[01/06/2023 14:50:28] Checked class coordinates
[01/06/2023 14:50:28] Cached version found, loading
[01/06/2023 14:50:28] Loading filtered classes from /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/segmentation/classes.csv
[01/06/2023 14:50:28] Loaded 485 filtered classes
[01/06/2023 14:50:28] After removing duplicates 485 filtered classes remain.
[01/06/2023 14:50:28] Extraction Details:
[01/06/2023 14:50:28] --------------------------------
[01/06/2023 14:50:28] Input channels: 3
[01/06/2023 14:50:28] Input labels: 2
[01/06/2023 14:50:28] Output channels: 5
[01/06/2023 14:50:28] Number of classes to extract: 484
[01/06/2023 14:50:28] Starting extraction of 484 classes
extracting classes: 16it [00:00, 157.74it/s]
[01/06/2023 14:50:28] Extracting dataset 0
extracting classes: 122it [00:00, 160.05it/s]
[01/06/2023 14:50:29] Extracting dataset 100
extracting classes: 234it [00:01, 177.51it/s]
[01/06/2023 14:50:29] Extracting dataset 200
extracting classes: 328it [00:01, 169.79it/s]
[01/06/2023 14:50:30] Extracting dataset 300
extracting classes: 422it [00:02, 180.15it/s]
[01/06/2023 14:50:30] Extracting dataset 400
extracting classes: 484it [00:02, 172.14it/s]
[01/06/2023 14:50:31] Finished extraction in 2.81 seconds (172.07 cells / second)
[01/06/2023 14:50:31] Collect cells
[01/06/2023 14:50:31] number of cells too close to image edges to extract: 52
[01/06/2023 14:50:31] Transferring extracted single cells to .hdf5
[01/06/2023 14:50:31] index created.
[01/06/2023 14:50:32] Tempmmap Folder location /var/folders/35/p4c58_4n3bb0bxnzgns1t7kh0000gn/T/./temp_mmap24gm079o will now be removed.
[01/06/2023 14:50:32] Finished cleaning up cache
Look at extracted single-cell images
The extracted single-cell images are written to a h5py file single_cells.h5 located under extraction\data within the project folder.
The file contains two datasets: single_cell_data and single_cell_index. single_cell_data contains the extracted single cell images while single_cell_index contains the cell id of the extracted cell at that location.
[51]:
import h5py
with h5py.File(f"{project_location}/extraction/data/single_cells.h5") as hf:
print(hf.keys())
<KeysViewHDF5 ['single_cell_data', 'single_cell_index']>
So if we want to look at the nth cell in the dataset we can first check which cellid was assigned to this cell by looking at the nth entry in the single_cell_index dataset and then get the extracted single-cell images from the single_cell_data dataset.
The extracted single-cell images represent the following information in this order:
nucleus mask 2. cytosol mask 3. nucleus channel 4. cytosol channel 5. other channels
Here we will demonstrate with the 10th cell in the dataset.
[52]:
with h5py.File(f"{project_location}/extraction/data/single_cells.h5") as hf:
index = hf.get("single_cell_index")
images = hf.get("single_cell_data")
print("cell id: ", index[10])
image = images[10]
fig, axs = plt.subplots(1, 5)
for i, _img in enumerate(image):
axs[i].imshow(_img)
axs[i].axis("off")
cell id: [10 11]
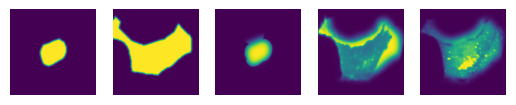
Classification of extracted single-cells
Next we can apply a pretained model to classify our cells within the SPARCSpy project.
Within the config.yml we specify which model should be used for inference and we can give it a name.
MLClusterClassifier:
channel_classification: 4
threads: 24 #
batch_size: 900
dataloader_worker: 0 #needs to be 0 if using cpu
standard_scale: False
exp_transform: False
log_transform: False
network: "autophagy_classifier2.1"
classifier_architecture: "VGG2_old"
screen_label: "Autophagy_15h_classifier2.1"
epoch: "max"
encoders: ["forward"]
inference_device: "cpu"
Here e.g. we are using a pretrained model included within the SPARCSpy library autophagy_classifier2.1 and are naming the results from this model Autophagy_15h_classifier2.1.
Model overview:
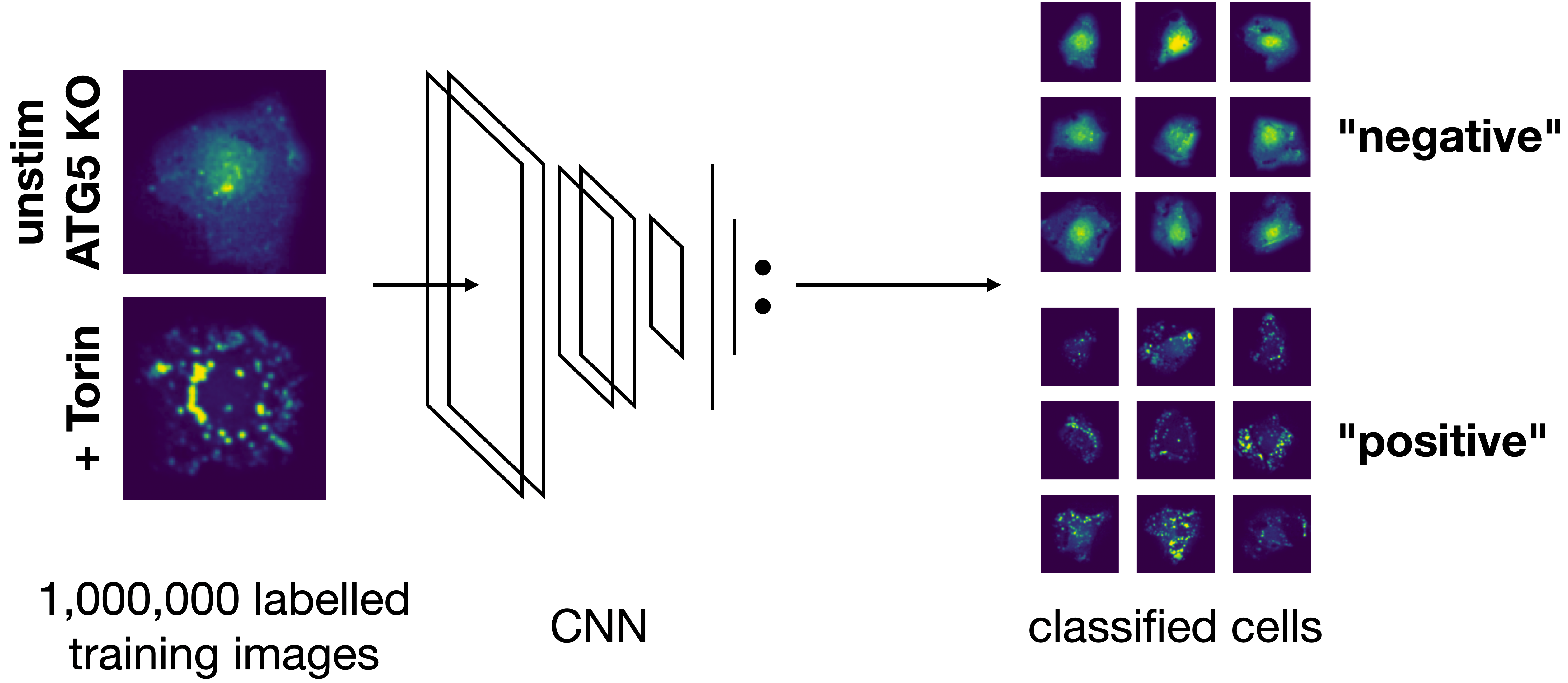
The inference results will be written to a new folder generated under classification with this name.
If we want to use a model we trained ourselves that is not yet included within the SPARCSpy library we can simply replace the network name in the config with the path to the checkpoint file generated by pytorch.
[53]:
accessory = ([], [], [])
project.classify(accessory = accessory)
Lightning automatically upgraded your loaded checkpoint from v1.5.5 to v2.0.2. To apply the upgrade to your files permanently, run `python -m pytorch_lightning.utilities.upgrade_checkpoint --file ../../../src/pretrained_models/autophagy/autophagy2.2/VGG2_autophagy_classifier2_2.cpkt`
[01/06/2023 14:50:32] Started classification
[01/06/2023 14:50:32] starting with run 0
[01/06/2023 14:50:32] channel_classification: 4
[01/06/2023 14:50:32] threads: 24
[01/06/2023 14:50:32] batch_size: 900
[01/06/2023 14:50:32] dataloader_worker: 0
[01/06/2023 14:50:32] standard_scale: False
[01/06/2023 14:50:32] exp_transform: False
[01/06/2023 14:50:32] log_transform: False
[01/06/2023 14:50:32] pca_dimensions: 0
[01/06/2023 14:50:32] umap_neighbours: 30
[01/06/2023 14:50:32] umap_min_dist: 0.001
[01/06/2023 14:50:32] network: autophagy_classifier2.2
[01/06/2023 14:50:32] classifier_architecture: VGG2_old
[01/06/2023 14:50:32] screen_label: Autophagy_15h_classifier2.1
[01/06/2023 14:50:32] epoch: max
[01/06/2023 14:50:32] encoders: ['forward']
[01/06/2023 14:50:32] inference_device: cpu
[01/06/2023 14:50:32] 0 different accessory datasets specified
[01/06/2023 14:50:32] model parameters loaded from autophagy_classifier2.2
[01/06/2023 14:50:32] loading /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/extraction/data
[01/06/2023 14:50:32] Total: 432
[01/06/2023 14:50:32] 0: 432
[01/06/2023 14:50:32]
[01/06/2023 14:50:32] log transfrom: False
[01/06/2023 14:50:32] start processing 1 batches with forward based inference
[01/06/2023 14:50:36] finished processing
looking at the generated results
The results are written to a csv file which we can load with pandas.
SPARCSpy writes the softmax results directly to csv as ln() for better precision. To transform these numbers back to the range between 0 and 1 we first need to apply the np.exp function.
[54]:
results = pd.read_csv(f"{project_location}/classification/0_Autophagy_15h_classifier2.1/dimension_reduction_forward.tsv", index_col = 0)
results.result_0 = np.exp(results.result_0)
results.result_1 = np.exp(results.result_1)
results
[54]:
| result_0 | result_1 | label | cell_id | |
|---|---|---|---|---|
| 0 | 9.229987e-07 | 0.999999 | 0 | 98 |
| 1 | 1.391613e-06 | 0.999999 | 0 | 60 |
| 2 | 2.630094e-05 | 0.999974 | 0 | 3 |
| 3 | 6.641098e-04 | 0.999336 | 0 | 103 |
| 4 | 1.444993e-02 | 0.985550 | 0 | 113 |
| ... | ... | ... | ... | ... |
| 427 | 3.525092e-05 | 0.999965 | 0 | 282 |
| 428 | 9.838445e-04 | 0.999016 | 0 | 423 |
| 429 | 6.322246e-06 | 0.999994 | 0 | 203 |
| 430 | 2.172819e-04 | 0.999783 | 0 | 114 |
| 431 | 9.377195e-07 | 0.999999 | 0 | 434 |
432 rows × 4 columns
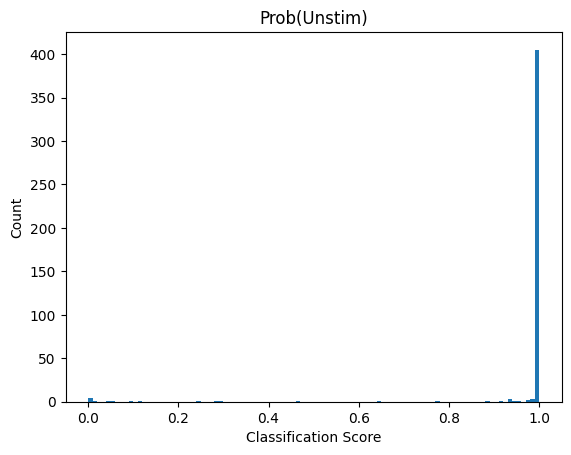
Depending on the model we used what result_0 and result_1 represent will vary. Here we used a binary classification model were class 1 was cells deficient in autophagy. So result_1 indicates the probability score that a cell has the label “autophagy off”. results_0 is simply 1 - result_1
If we look at the distribution in our dataset we can see that almost all cells are classified as “autophagy defective”. Since the input images were from unstimulated wt cells this matches to our expectation.
[55]:
plt.hist(results.result_1, bins = 100);
plt.title("Prob(Unstim)");
plt.xlabel("Classification Score");
plt.ylabel("Count");
[ ]:
Exporting Cutting contours for excision on the LMD7
SPARCSpy directly interfaces with our other open-source python library py-lmd to easily select and export cells for excision on a Leica LMD microscope.
Of note: this will require that the cells have been plates on a LMD compatible surface (like a PPS slide). SPARCSpy can of course also simply be used for data analysis procedures, then ignore this last step.
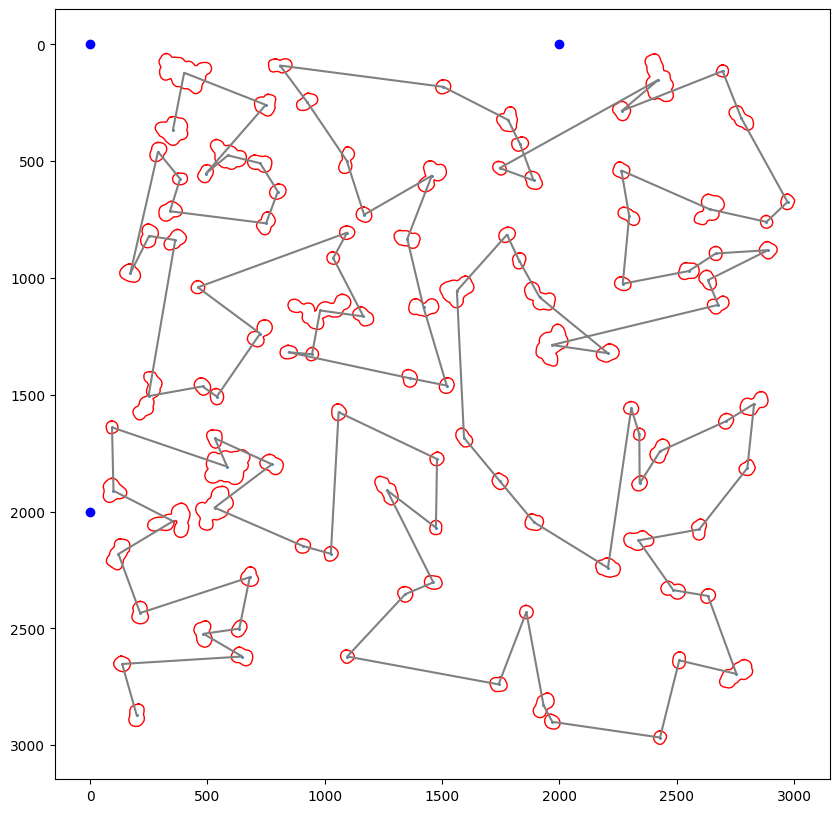
First we will select the cells we wish to excise based on their classification score. Here we have chosen a threadhold >= 0.99999 for bin1 and a threshold <= 0.9 for bin2.
[56]:
cell_ids_bin1 = results[results.result_1 >= 0.99999].cell_id.tolist()
cell_ids_bin2 = results[results.result_1 <= 0.9].cell_id.tolist()
print("number of cells to excise:",len(cell_ids_bin1) + len(cell_ids_bin2))
number of cells to excise: 271
The cells can then be allocated into different wells.
[57]:
cells_to_select = [{"name": "bin1", "classes": list(cell_ids_bin1), "well":"A1"},
{"name": "bin2", "classes": list(cell_ids_bin2), "well":"B1"},
]
In addition to defining which cells we want to excise, we also need to pass the location of the calibration crosses so that we can transfer the image coordinate system into a cutting coordinate system. You can read up on this here.
To obtain the coordinates of your reference points simply open your stitched image in e.g. FIJI and navigate to the location of your reference points. Then write out the coordinates for each point.
[58]:
marker_0 = (0, 0)
marker_1 = (2000, 0)
marker_2 = (0, 2000)
calibration_marker = np.array([marker_0, marker_1, marker_2])
As with the previous methods, additional parameters can be passed to the selection function via the config.yml file which adapts the behaviour of how cutting shapes are generated. You can read up more on this here.
LMDSelection:
processes: 20
# defines the channel used for generating cutting masks
# segmentation.hdf5 => labels => segmentation_channel
# When using WGA segmentation:
# 0 corresponds to nuclear masks
# 1 corresponds to cytosolic masks.
segmentation_channel: 0
# dilation of the cutting mask in pixel
shape_dilation: 16
# number of datapoints which are averaged for smoothing
# the number of datapoints over an distance of n pixel is 2*n
smoothing_filter_size: 25
# fold reduction of datapoints for compression
poly_compression_factor: 30
# can be "none", "greedy", or "hilbert"
path_optimization: "hilbert"
# number of nearest neighbours for optimized greedy search
greedy_k: 15
# hilbert curve order
hilbert_p: 7
[59]:
project.select(cells_to_select, calibration_marker, debug=True)
[01/06/2023 14:50:37] Selection process started
No configuration for shape_erosion found, parameter will be set to 0
No configuration for binary_smoothing found, parameter will be set to 3
No configuration for convolution_smoothing found, parameter will be set to 15
No configuration for xml_decimal_transform found, parameter will be set to 100
No configuration for distance_heuristic found, parameter will be set to 300
No configuration for join_intersecting found, parameter will be set to True
sanity check for cell set 0
cell set 0 passed sanity check
sanity check for cell set 1
cell set 1 passed sanity check
Convert label format into coordinate format
Conversion finished, sanity check
Check passed
Check passed
Check passed
100%|██████████| 255/255 [00:02<00:00, 115.79it/s]
Create shapes for merged cells
100%|██████████| 106/106 [00:01<00:00, 57.49it/s]
Calculating polygons
100%|██████████| 106/106 [00:03<00:00, 31.44it/s]
Polygon calculation finished
Current path length: 142,403.15 units
Path optimizer defined in config: hilbert
Optimized path length: 142,403.15 units
Optimization factor: 1.0x
Convert label format into coordinate format
Conversion finished, sanity check
Check passed
Check passed
Check passed
100%|██████████| 16/16 [00:01<00:00, 10.87it/s]
Create shapes for merged cells
100%|██████████| 15/15 [00:01<00:00, 10.66it/s]
Calculating polygons
100%|██████████| 15/15 [00:01<00:00, 10.55it/s]
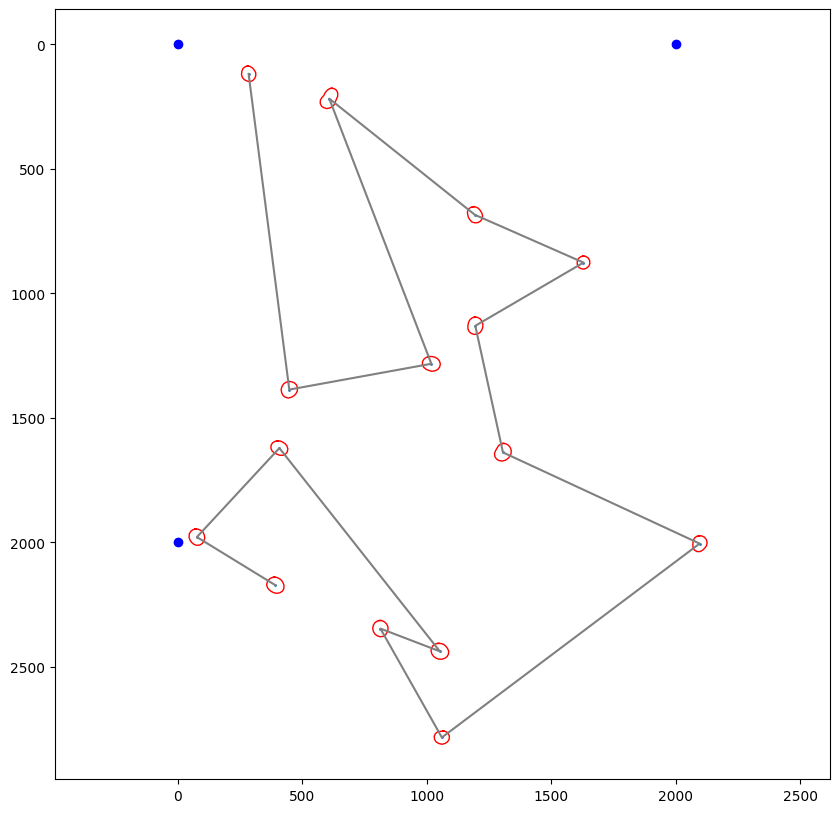
Polygon calculation finished
Current path length: 17,031.39 units
Path optimizer defined in config: hilbert
Optimized path length: 17,031.39 units
Optimization factor: 1.0x
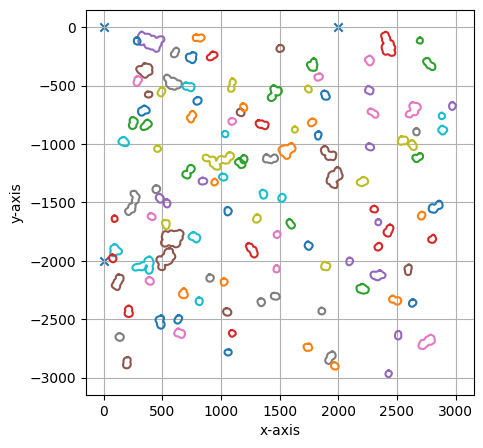
===== Collection Stats =====
Number of shapes: 121
Number of vertices: 2,912
============================
Mean vertices: 24
Min vertices: 14
5% percentile vertices: 16
Median vertices: 20
95% percentile vertices: 50
Max vertices: 70
[0 0]
[ 0 -200000]
[200000 0]
[01/06/2023 14:50:52] Saved output at /Users/sophia/Documents/GitHub/SPARCSpy/example_data/example_1/project/selection/bin1_bin2.xml