4.1. lib#
- class lmd.lib.Collection(calibration_points: ndarray | None = None, orientation_transform: ndarray | None = None, scale: float = 100)#
Class which is used for creating shape collections for the Leica LMD6 & 7. Contains a coordinate system defined by calibration points and a collection of various shapes.
- Parameters:
calibration_points – Calibration coordinates in the form of \((3, 2)\).
orientation_transform – defines transformations performed on the provided coordinate system prior to export as XML. Defaults to the identity matrix.
- calibration_points#
Calibration coordinates in the form of \((3, 2)\).
- Type:
Optional[np.ndarray]
- orientation_transform#
defines transformations performed on the provided coordinate system prior to export as XML. This orientation_transform is always applied to shapes when there is no individual orientation_transform provided.
- Type:
np.ndarray
- add_shape(shape: Shape)#
Add a new shape to the collection.
- Parameters:
shape – Shape which should be added.
- join(collection: Collection, update_orientation_transform: bool = True)#
Join the collection with the shapes of a different collection. The calibration markers of the current collection are kept. Please keep in mind that coordinate systems and calibration points must be compatible for correct joining of collections.
- Parameters:
collection – Collection which should be joined with the current collection object.
orientation_transform – If set to True, the orientation transform of the joined collection will be updated to the current collection. If set to False, the orientation transform of the joined collection will not be updated.
- Returns:
returns self
- load(file_location: str, *, raise_shape_errors: bool = False)#
Can be used to load a shape file from XML. Both, XMLs generated with py-lmd and the Leica software can be used. :param file_location: File path pointing to the XML file. :param raise_errors: Whether to raise errors during shape collection. If False raises a warning.
- load_geopandas(gdf: GeoDataFrame, geometry_column: str = 'geometry', name_column: str | None = None, well_column: str | None = None, calibration_points: ndarray | None = None, global_coordinates: int | None = None, custom_attribute_columns: str | list[str] | None = None) None#
Create collection from a geopandas dataframe
- Parameters:
gdf (geopandas.GeoDataFrame) – Collection of shapes and optional metadata
(str (geometry_column) – geometry): Name of column storing Shapes as shapely.Polygon, defaults to geometry
default – geometry): Name of column storing Shapes as shapely.Polygon, defaults to geometry
well_column (str, optional) – Column storing of well id as additional metadata
calibration_points (np.ndarray, optional) – Calibration points of collection
global_coordinates (int, optional) – Number of global coordinates
shape. (custom_attribute_columns Custom shape metadata that will be added as additional xml-element to the) – Can be column name, list of column names or None
Example:
from lmd.lib import Collection import geopandas as gpd import shapely gdf = gpd.GeoDataFrame( data={"well": ["A1"], "name": ["test"]}, geometry=[shapely.Polygon([[0, 0], [0, 1], [1, 0], [0, 0]])] ) # Create collection c = Collection() # Export well metadata c.load_geopandas(gdf, well_column="well") assert c.to_geopandas("well").equals(gdf) # Do not export well metadata c.load_geopandas(gdf) assert c.to_geopandas().equals(gdf.drop(columns="well"))
- new_shape(points: ndarray, well: str | None = None, name: str | None = None, **custom_attributes)#
Directly create a new Shape in the current collection.
- Parameters:
points – Array or list of lists in the shape of (N,2). Contains the points of the polygon forming a shape.
well – Well in which to sort the shape after cutting. For example A1, A2 or B3.
name – Name of the shape.
custom_attributes – Custom shape metadata that can be added as additional xml-element to the shape. All values are converted to strings.
- plot(calibration: bool = True, mode: str = 'line', fig_size: tuple = (5, 5), apply_orientation_transform: bool = True, apply_scale: bool = False, save_name: str | None = None, return_fig: bool = False, **kwargs)#
This function can be used to plot all shapes of the corresponding shape collection.
- Parameters:
calibration – Controls wether the calibration points should be plotted as crosshairs. Deactivating the crosshairs will result in the size of the canvas adapting to the shapes. Can be especially usefull for small shapes or debugging.
fig_size – Defaults to \((10, 10)\) Controls the size of the matplotlib figure. See matplotlib documentation for more information.
apply_orientation_transform – Define wether the orientation transform should be applied before plotting.
(Optional[str] (save_name) – None): Specify a filename for saving the generated figure. By default None is provided which will not save a figure.
default – None): Specify a filename for saving the generated figure. By default None is provided which will not save a figure.
- save(file_location: str, encoding: str = 'utf-8')#
Can be used to save the shape collection as XML file.
file_location: File path pointing to the XML file.
- stats()#
Print statistics about the Collection in the form of:
===== Collection Stats ===== Number of shapes: 208 Number of vertices: 126,812 ============================ Mean vertices: 609.67 Min vertices: 220.00 5% percentile vertices: 380.20 Median vertices: 594.00 95% percentile vertices: 893.20 Max vertices: 1,300.00
- svg_to_lmd(file_location, offset=None, divisor=3, multiplier=60, rotation_matrix=array([[1., 0.], [0., 1.]]), orientation_transform=None)#
Can be used to save the shape collection as XML file.
- Parameters:
file_location – File path pointing to the SVG file.
orientation_transform – Will superseed the global transform of the Collection.
rotation_matrix
- to_geopandas(*attrs: str) GeoDataFrame#
Return geopandas dataframe of collection
- Parameters:
*attrs (str) – Optional attributes of the shapes in the collection to be added as metadata columns
- Returns:
Representation of all shapes and optional metadata
- Return type:
geopandas.GeoDataFrame
Example: .. code-block:: python
# Generate collection collection = pylmd.Collection() shape = pylmd.Shape(
np.array([[ 0, 0], [ 0, -1], [ 1, 0], [ 0, 0]]), well=”A1”, name=”Shape_1”, metadata1=”A”, metadata2=”B”, orientation_transform=None
)
collection.add_shape(shape)
# Get geopandas object collection.to_geopandas() > geometry
0 POLYGON ((0 0, 0 -1, 1 0, 0 0))
collection.to_geopandas(“well”, “name”, “metadata1”, “metadata2”) > well name metadata1 metadata2 geometry
0 A1 Shape_1 A B POLYGON ((0 0, 0 -1, 1 0, 0 0))
- class lmd.lib.SegmentationLoader(config=None, verbose=False, processes=1)#
Select single cells from a segmentation and generate cutting data
- Args:
config (dict): Dict containing configuration parameters. See Note for further explanation. processes (int): Number of processes used for parallel processing of cell sets. Total processes can be calculated as processes * threads. threads (int): Number of threads used for parallel processing of shapes within a cell set. Total processes can be calculated as processes * threads.
cell_sets (list(dict)): List of dictionaries containing the sets of cells which should be sorted into a single well.
calibration_marker (np.array): Array of size ‘(3,2)’ containing the calibration marker coordinates in the ‘(row, column)’ format.
coords_lookup (None, dict): precalculated lookup table for coordinates of individual cell ids. If not provided will be calculated.
classes (np.array): Array of classes found in the provided segmentation mask. If not provided will be calculated based on the assumption that cell_ids are assigned in ascending order.
Example:
import numpy as np from PIL import Image from lmd.lib import SegmentationLoader
- Parameters:
config (dict) – Dict containing configuration parameters. See Note for further explanation.
cell_sets (list(dict)) – List of dictionaries containing the sets of cells which should be sorted into a single well.
calibration_marker (np.array) – Array of size ‘(3,2)’ containing the calibration marker coordinates in the ‘(row, column)’ format.
Example
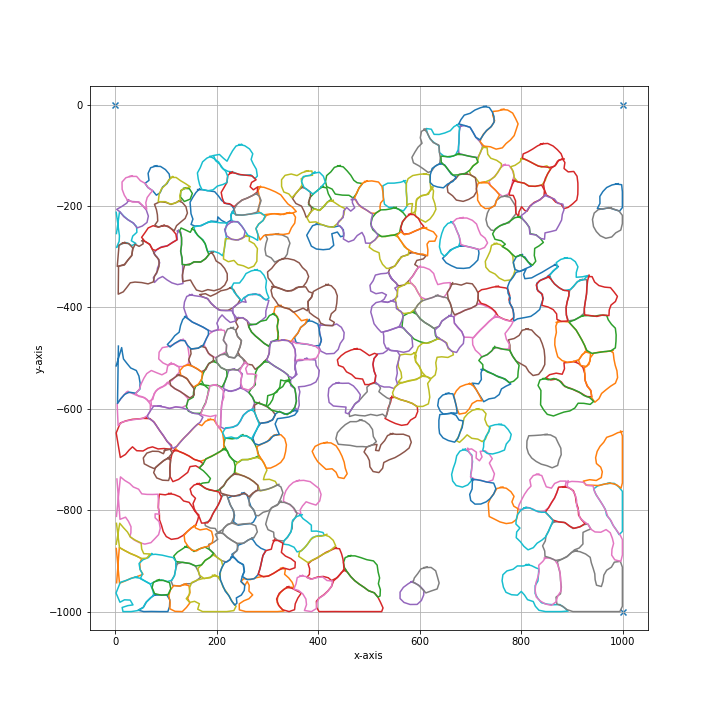
import numpy as np from PIL import Image from lmd.lib import SegmentationLoader from lmd._utils import _download_segmentation_example_file # use example image provided within py-lmd example_image_path = _download_segmentation_example_file() im = Image.open(example_image_path) segmentation = np.array(im).astype(np.uint32) all_classes = np.unique(segmentation) cell_sets = [{"classes": all_classes, "well": "A1"}] calibration_points = np.array([[0, 0], [0, 1000], [1000, 1000]]) loader_config = {"orientation_transform": np.array([[0, -1], [1, 0]])} sl = SegmentationLoader(config=loader_config) shape_collection = sl(segmentation, cell_sets, calibration_points) shape_collection.plot(fig_size=(10, 10))
Note
Basic explanation of the parameters in the config dict:
# dilation of the cutting mask in pixel before intersecting shapes in a selection group are merged shape_dilation: 0 # erosion of the cutting mask in pixel before intersecting shapes in a selection group are merged shape_erosion: 0 # Cutting masks are transformed by binary dilation and erosion binary_smoothing: 3 # number of datapoints which are averaged for smoothing # the resoltion of datapoints is twice as high as the resolution of pixel convolution_smoothing: 15 # strength of coordinate reduction through the Ramer-Douglas-Peucker algorithm 0 is small 1 is very high rdp_epsilon: 0.1 # Optimization of the cutting path inbetween shapes # optimized paths improve the cutting time and the microscopes focus # valid options are ["none", "hilbert", "greedy"] path_optimization: "hilbert" # Paramter required for hilbert curve based path optimization. # Defines the order of the hilbert curve used, which needs to be tuned with the total cutting area. # For areas of 1 x 1 mm we recommend at least p = 4, for whole slides we recommend p = 7. hilbert_p: 7 # Parameter required for greedy path optimization. # Instead of a global distance matrix, the k nearest neighbours are approximated. # The optimization problem is then greedily solved for the known set of nearest neighbours until the first set of neighbours is exhausted. # Established edges are then removed and the nearest neighbour approximation is recursivly repeated. greedy_k: 20 # Overlapping shapes are merged based on a nearest neighbour heuristic. # All selected shapes closer than distance_heuristic pixel are checked for overlap. distance_heuristic: 300
- DEFAULT_SEGMENTATION_DTYPE#
alias of
uint64
- check_cell_set_sanity(cell_set)#
Check if cell_set dictionary contains the right keys
- load_classes(cell_set)#
Identify cell class definition and load classes
Identify if cell classes are provided as list of integers or as path pointing to a csv file. Depending on the type of the cell set, the classes are loaded and returned for selection.
- class lmd.lib.Shape(points: ndarray = array([[5.e-324, 0.e+000]]), well: str | None = None, name: str | None = None, orientation_transform=None, **custom_attributes: dict[str, str])#
Class for creating a single shape object.
- classmethod from_xml(root, orientation_transform: ndarray | None = None)#
Load a shape from an XML shape node. Used internally for reading LMD generated XML files.
- Parameters:
root – XML input node.
- get_shape_annotation(name: str) Any | None#
- Retrieve the value of an attribute from either instance attributes
or custom attributes by name.
Searches for the attribute by name in the 1) instance attributes 2) custom attributes, or 3) issues a warning and returns None
- Parameters:
name (str) – The name of the attribute to retrieve.
- Returns:
The value of the attribute if found, otherwise None.
- Return type:
Any | None
- to_xml(id: int, orientation_transform: ndarray, scale: float, *, write_custom_attributes: bool = True)#
Generate XML shape node needed internally for export.
- Parameters:
id – Sequential identifier of the shape as used in the LMD XML format.
orientation_transform (np.array) – Pass orientation_transform which is used if no local orientation transform is set.
scale (float) – Scalling factor used to enable higher decimal precision.
write_custom_attributes – Write custom attributes to xml file
Note
If the Shape has a custom orientation_transform defined, the custom orientation_transform is applied at this point. If not, the oritenation_transform passed by the parent Collection is used. This highlights an important difference between the Shape and Collection class. The Collection will always has an orientation transform defined and will use np.eye(2) by default. The Shape object can have a orientation_transform but can also be set to None to use the Collection value.