3.4. Generate Calibration Cross Templates for PPS membrane slides#
This calibration template contains 3 larger calibration crosses in the top left corner, the bottom left corner and the middle of the right side as well as a slide ID number.
[1]:
import numpy as np
import os
from lmd.lib import Collection
from lmd.tools import text, makeCross
[ ]:
# ============ parameters ============
# these calibration points are compatible with the calibration marker preset DOES NOT NEED TO BE CHANGED FOR REGULAR PPS MEMBRANE SLIDES
n_marker = np.array([[0, 27610], [0, 0], [9690, 0]])
# offset of the raster in um
offset = np.array([1000, 1000])
# size of a single unit cell
# the margin is subtracted from this to give the final shape
size = 324 * 0.25
# margin as fraction of the unit cell size
margin = 0.2
# number of rows/columns
N = 100
# 90 degree clockwise. DOES NOT NEED TO BE CHANGED
orientation_transform = np.array([[-1, 0], [0, -1]])
# =====================================
size_px = size / 1.62
offset_px = offset / 1.62
# =====================================
# define ID number to be burned onto slide
id = 1000
outdir = "../../../src/pylmd_data/calibration_templates"
if not os.path.exists(outdir):
os.makedirs(outdir)
[3]:
shapefile = Collection()
shapefile.calibration_points = n_marker
offset = np.array([500, 27110])
offset_letter = offset + np.array([-100, 100])
cross1 = makeCross(offset, [50, 100, 100, 50], 0.5, 10)
shapefile.join(cross1)
shapefile.join(text("A", offset=offset_letter, multiplier=7))
offset = np.array([500, 500])
offset_letter = offset + np.array([-100, -100])
cross2 = makeCross(offset, [100, 100, 50, 50], 0.5, 10)
shapefile.join(cross2)
shapefile.join(text("B", offset=offset_letter, multiplier=7))
offset = np.array([9100, 13500])
offset_letter = offset + np.array([100, 100])
cross3 = makeCross(offset, [100, 50, 100, 100], 0.5, 10)
shapefile.join(cross3)
shapefile.join(text("C", offset=offset_letter, multiplier=7))
# generate slide ID number
offset_ID = np.array([800, 27110])
shapefile.join(text(str(id).zfill(3), offset=offset_ID, multiplier=7))
shapefile.plot(calibration=True, fig_size=(10, 30))
shapefile.save(f"{outdir}/calibration_template_{id}.xml")
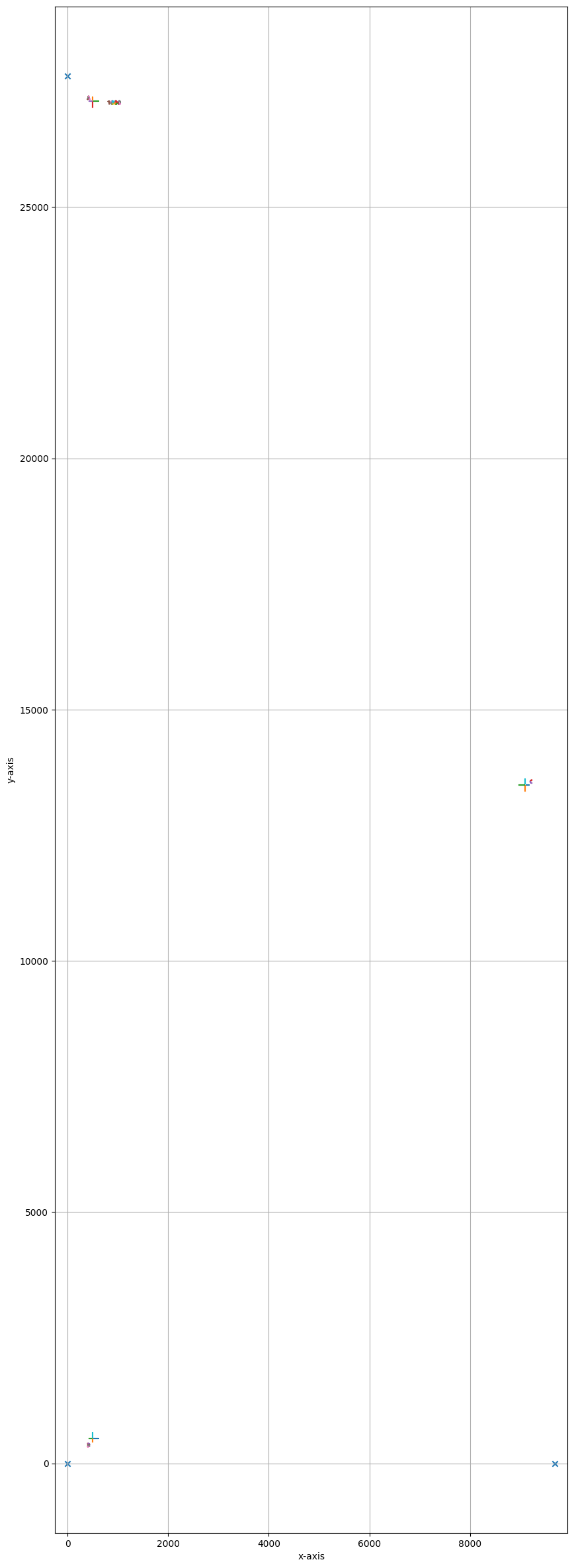
[ 0. 2761000.]
[0. 0.]
[969000. 0.]