3.2. Generate cutting XML from exported results from QUPath#
The stitched images were loaded into Qupath and specific regions annotaed by hand. calibration points were also selected and labelled with calib1, calib2 and calib3.
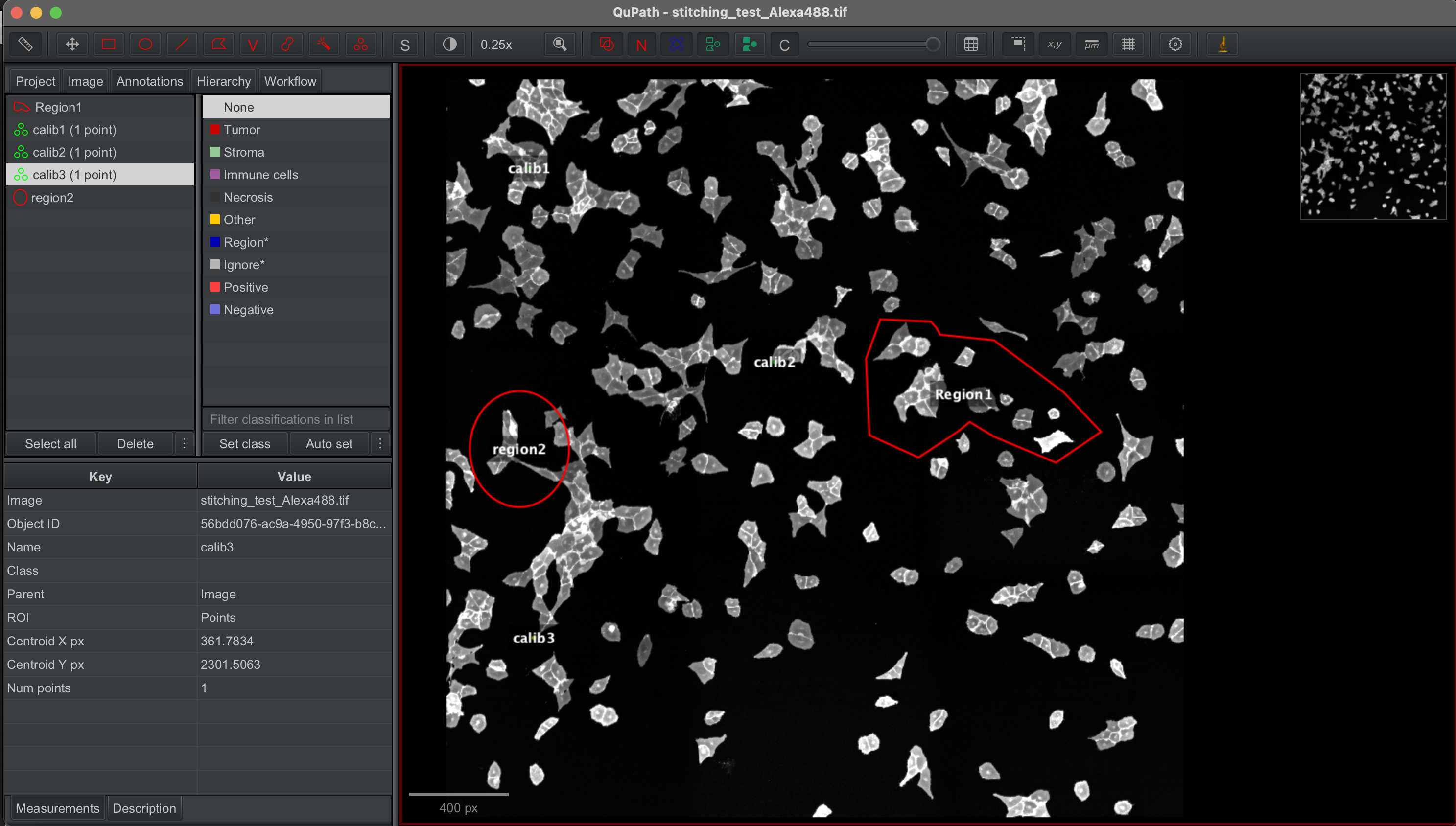
The annotated shapes were then exported to geojson file.
3.2.1. Import libraries and define helper functions#
[1]:
import shapely
import geopandas
import pandas as pd
import numpy as np
from lmd.lib import Collection
[2]:
def separate_points_polygons(
df: geopandas.GeoDataFrame, geometry_column: str = "geometry"
) -> tuple[geopandas.GeoDataFrame, geopandas.GeoDataFrame]:
"""Split QuPath geopandas dataframe by shape types"""
points = df[geometry_column].apply(lambda geom: isinstance(geom, shapely.Point))
polygons = df[geometry_column].apply(lambda geom: isinstance(geom, shapely.Polygon))
return df.loc[points], df.loc[polygons]
[3]:
def get_calib_points(list_of_calibpoint_names: str, df: geopandas.GeoDataFrame) -> np.ndarray:
"""Parse selected points into np.array"""
# Create list of relevant shapes
pointlist = []
for point_name in list_of_calibpoint_names:
pointlist.append(df.loc[df["name"] == point_name, "geometry"].values[0])
return np.array([[point.x, point.y] for point in pointlist])
3.2.2. Import GEOjson regions#
The geojson dataset is loaded into a dataframe with the following structure:
id |
objectType |
classification |
name |
geometry |
|---|---|---|---|---|
unique shape id |
type of shape (e.g. annotation) |
all annotation information from qupath |
shape name if given |
contains information relevant for shape |
The geojson should besides containing individual segemnted shapes contain 3 points annotated as calib1, calib2, calib3 that will be used as calibration points for generating the XML
[4]:
df = geopandas.read_file("test_data/cellculture_example/annotated_regions_Qupath.geojson")
df
Skipping field color: unsupported OGR type: 1
[4]:
| id | objectType | name | geometry | |
|---|---|---|---|---|
| 0 | 9287277d-e46f-47e9-aa3f-c540b3318b5e | annotation | region2 | POLYGON ((507 1524, 506.6 1539.01, 505.39 1553... |
| 1 | ab63cfd3-17d0-4dd3-b8e6-95172dda64de | annotation | Region1 | POLYGON ((1789 990, 1730 1153, 1744 1467, 1944... |
| 2 | b60d2cf7-963e-42ae-8a2a-69ff289d29db | annotation | calib1 | POINT (343.24 368.53) |
| 3 | 56bdd076-ac9a-4950-97f3-b8c2268ee090 | annotation | calib3 | POINT (361.78 2301.51) |
| 4 | 03d9bb6e-16b1-4cd9-9181-86da90fb98bf | annotation | calib2 | POINT (1353.77 1165.83) |
Annotations and calibration points (shapely.Point objects) and annoatations (shapely.Polygon objects) should be processed separately. Therefore, we will use the previously defined helper function separate_points_polygons to split the dataframe by shape type.
[5]:
# Separate calibration points (shapely.Points) from annotations (shapely.Polygons)
points, annotations = separate_points_polygons(df)
Let’s inspect the separated geodataframes
[6]:
points
[6]:
| id | objectType | name | geometry | |
|---|---|---|---|---|
| 2 | b60d2cf7-963e-42ae-8a2a-69ff289d29db | annotation | calib1 | POINT (343.24 368.53) |
| 3 | 56bdd076-ac9a-4950-97f3-b8c2268ee090 | annotation | calib3 | POINT (361.78 2301.51) |
| 4 | 03d9bb6e-16b1-4cd9-9181-86da90fb98bf | annotation | calib2 | POINT (1353.77 1165.83) |
[7]:
annotations
[7]:
| id | objectType | name | geometry | |
|---|---|---|---|---|
| 0 | 9287277d-e46f-47e9-aa3f-c540b3318b5e | annotation | region2 | POLYGON ((507 1524, 506.6 1539.01, 505.39 1553... |
| 1 | ab63cfd3-17d0-4dd3-b8e6-95172dda64de | annotation | Region1 | POLYGON ((1789 990, 1730 1153, 1744 1467, 1944... |
3.2.3. Calibration points#
pyLMD expects calibration points as numpy array of the shape (N, 2). We therefore need to extract the coordinates of the calibration points from the points geodataframe. To do so, we will use the second helper function get_calib_points that converts the geometry column of the selected points to a numpy array. Each row in the numpy array represents the coordinates of the calibration points in (x, y) format.
[8]:
caliblist = get_calib_points(["calib1", "calib2", "calib3"], points)
caliblist
[8]:
array([[ 343.24, 368.53],
[1353.77, 1165.83],
[ 361.78, 2301.51]])
3.2.4. Generate shape collection#
We are now able to convert the annotation dataframe to a collection of shapes. To do so, we initialize an empty pylmd Collection with the previously defined calibration points. We will further define the flip the axes of the points with an orientation_transform as the image coordinates are flipped compared to the microscope coordinates (for a detailed explanation see this tutorial)
[9]:
annotations
[9]:
| id | objectType | name | geometry | |
|---|---|---|---|---|
| 0 | 9287277d-e46f-47e9-aa3f-c540b3318b5e | annotation | region2 | POLYGON ((507 1524, 506.6 1539.01, 505.39 1553... |
| 1 | ab63cfd3-17d0-4dd3-b8e6-95172dda64de | annotation | Region1 | POLYGON ((1789 990, 1730 1153, 1744 1467, 1944... |
[10]:
orientation_transform = np.array([[1, 0], [0, -1]])
shape_collection = Collection(calibration_points=caliblist, orientation_transform=orientation_transform)
# Load geopandas into collection class
shape_collection.load_geopandas(annotations, name_column="name")
shape_collection.stats()
===== Collection Stats =====
Number of shapes: 2
Number of vertices: 117
============================
Mean vertices: 58
Min vertices: 16
5% percentile vertices: 20
Median vertices: 58
95% percentile vertices: 97
Max vertices: 101
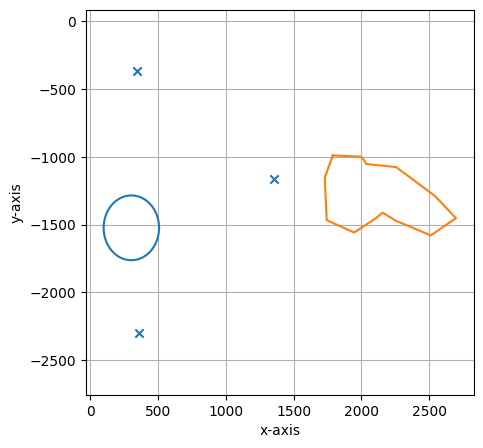
Let’s inspect the loaded annotations and calibration points with the internal plot function:
[11]:
shape_collection.plot(calibration=True)
3.2.5. Write to XML#
You can now write the selection to a Leica-compatible xml file
[12]:
shape_collection.save("./test_data/cellculture_example/shapes_2.xml")
[ 34324. -36853.]
[ 135377. -116583.]
[ 36178. -230151.]