Example Stitching Notebook
[1]:
import os
os.environ['JAVA_HOME'] = "/Users/sophia/mambaforge/envs/scPortrait/lib/jvm"
import matplotlib.pyplot as plt
from scportrait.tools.stitch import Stitcher, ParallelStitcher
/Users/sophia/mambaforge/envs/scPortrait/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Single-threaded Stitching
initializing the stitcher object
[2]:
input_dir = os.path.join("../../../../example_data/example_stitching/raw_image_tiles")
slidename = "stitching_test"
outdir = os.path.abspath(os.path.join("../../../../example_data/example_stitching/", slidename))
row = str(2).zfill(2) #specify the row of the well you want to stitch
well = str(4).zfill(2) #specifc the well number you wish to stitch
zstack_value = str(1).zfill(3) #specify the zstack you want to stitch. for multiple zstacks please make a loop and iterate through each of them.
timepoint = str(1).zfill(3) #specifz the timepoint you wish to stitch
pattern = f"Timepoint{timepoint}_Row{row}_Well{well}_{{channel}}_zstack{zstack_value}_r{{row:03}}_c{{col:03}}.tif"
#initialize stitcher
stitcher = Stitcher(os.path.abspath(input_dir),
slidename,
outdir,
pattern = pattern,
stitching_channel = "Alexa488",
overlap= 0.1,
max_shift= 30,
filter_sigma = 0,
rescale_range = {"Alexa488": (1, 99), "DAPI": (1, 99), "mCherry": (1, 99)},
overwrite=True)
Output directory at /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test already exists, overwriting.
Generating thumbnails
[3]:
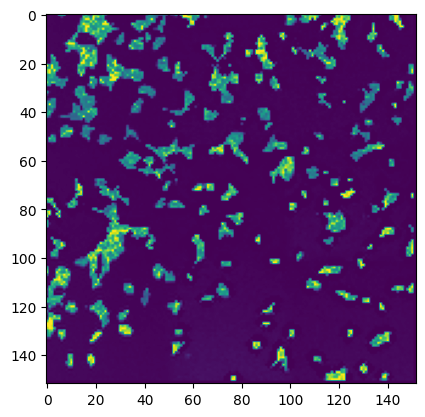
stitcher.generate_thumbnail()
assembling thumbnail 9/9
[4]:
#thumbnail is saved in the stitcher object and can be accessed via stitcher.thumbnail
plt.imshow(stitcher.thumbnail)
[4]:
<matplotlib.image.AxesImage at 0xa8e0f1ff0>
[5]:
#alterantively it can be saved to a tif file
stitcher.write_thumbnail()
[6]:
stitcher.initialize_reader()
Generating full-scale stitched image
[7]:
stitcher.stitch()
performing stitching on channel Alexa488 with id number 0
quantifying alignment error 1000/1000
aligning edge 12/12
WARNING:root:Folder /var/folders/35/p4c58_4n3bb0bxnzgns1t7kh0000gn/T/temp_mmap_eqb2txlq with temp mmap arrays is being deleted. All existing temp mmapp arrays will be unusable!
WARNING:root:New temp folder location. Temp mmap arrays are written to /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test/temp_mmap_qx6rbrpl. Cleanup of this folder is OS dependant, and might need to be triggered manually!
Alignment complete.
assembling mosaic with shape (3, 3040, 3038)
created tempmmap array for assembled mosaic at /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test/temp_mmap_qx6rbrpl/temp_mmap_8836257951842585069.hdf
0%| | 0/3 [00:00<?, ?it/s]
merging tile 9/9
33%|███▎ | 1/3 [00:01<00:02, 1.07s/it]
merging tile 9/9
67%|██████▋ | 2/3 [00:02<00:01, 1.07s/it]
merging tile 9/9
100%|██████████| 3/3 [00:03<00:00, 1.08s/it]
[8]:
#the stitched image is saved in the stitcher object and can be accessed via stitcher.assembled_mosaic
stitcher.assembled_mosaic
[8]:
|
||||||||||||||||
[9]:
# the stitched image can then be written to a variety of output formats
stitcher.write_tif(export_xml=True)
stitcher.write_ome_zarr()
[10]:
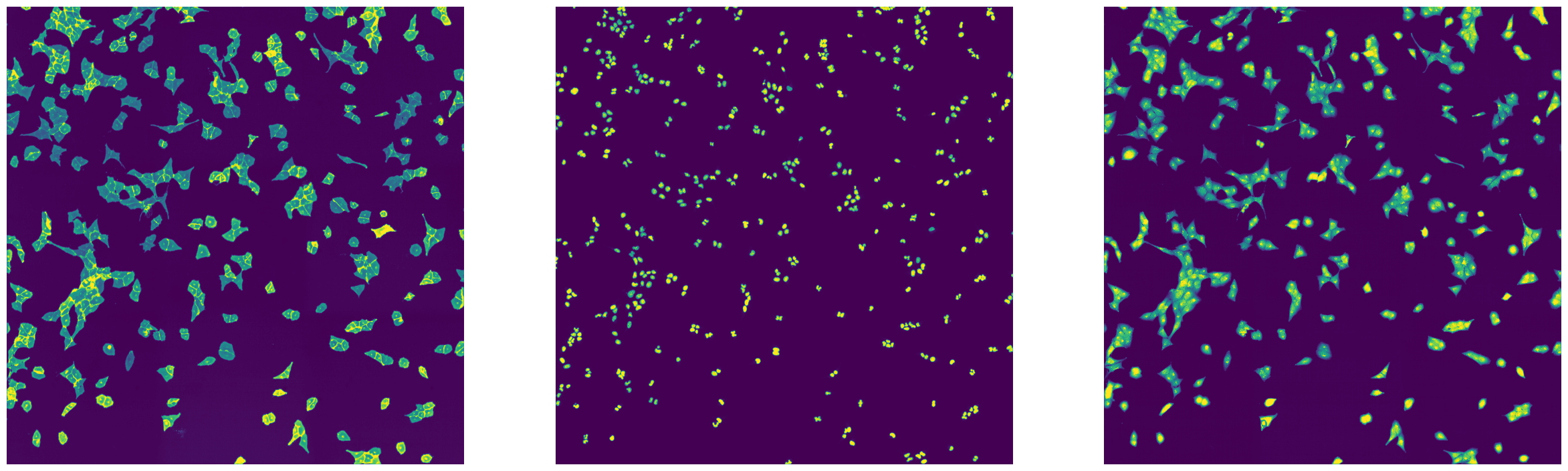
import matplotlib.pyplot as plt
from tifffile import imread
fig, axs = plt.subplots(1, 3, figsize = (30, 10))
axs[0].imshow(imread(f"{outdir}/stitching_test_Alexa488.tif"))
axs[0].axis("off")
axs[1].imshow(imread(f"{outdir}/stitching_test_DAPI.tif"))
axs[1].axis("off")
axs[2].imshow(imread(f"{outdir}/stitching_test_mCherry.tif"))
axs[2].axis("off")
[10]:
(-0.5, 3037.5, 3039.5, -0.5)
[11]:
del stitcher
Multi-threaded Stitching
Using the ParallelStitcher class stitching can be speed up by using multiple threads. The code to perform stitching remains more or less the same.
initializing the stitcher object
[12]:
input_dir = os.path.join("../../../../example_data/example_stitching/raw_image_tiles")
slidename = "stitching_test_parallel"
outdir_parallel = os.path.abspath(os.path.join("../../../../example_data/example_stitching/", slidename))
row = str(2).zfill(2) #specify the row of the well you want to stitch
well = str(4).zfill(2) #specifc the well number you wish to stitch
zstack_value = str(1).zfill(3) #specify the zstack you want to stitch. for multiple zstacks please make a loop and iterate through each of them.
timepoint = str(1).zfill(3) #specifz the timepoint you wish to stitch
pattern = f"Timepoint{timepoint}_Row{row}_Well{well}_{{channel}}_zstack{zstack_value}_r{{row:03}}_c{{col:03}}.tif"
#initialize stitcher
stitcher = ParallelStitcher(input_dir,
slidename,
outdir_parallel,
pattern = pattern,
stitching_channel = "Alexa488",
overlap= 0.1,
max_shift= 30,
filter_sigma = 0,
rescale_range = {"Alexa488": (1, 99), "DAPI": (1, 99), "mCherry": (1, 99)},
overwrite=True,
threads = 12)
Output directory at /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test_parallel already exists, overwriting.
Generating thumbnails
[13]:
stitcher.generate_thumbnail()
assembling thumbnail 1/9 assembling thumbnail 9/9
[14]:
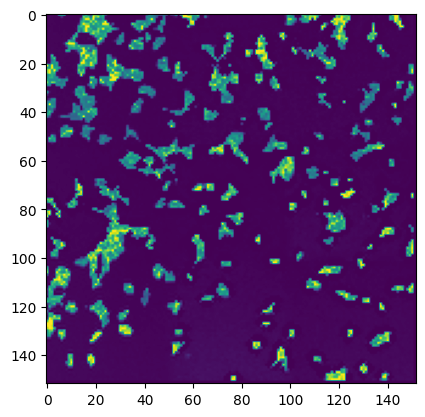
#thumbnail is saved in the stitcher object and can be accessed via stitcher.thumbnail
plt.imshow(stitcher.thumbnail)
[14]:
<matplotlib.image.AxesImage at 0xa9320ec20>
[15]:
#alterantively it can be saved to a tif file
stitcher.write_thumbnail()
Generating full-scale stitched image
[16]:
stitcher.stitch()
performing stitching on channel Alexa488 with id number 0
networkx
HERE
quantifying alignment error: 100%|██████████| 1000/1000 [00:00<00:00, 1187.36it/s]
aligning edge: 100%|██████████| 12/12 [00:00<00:00, 92.61it/s]
WARNING:root:Folder /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test/temp_mmap_qx6rbrpl with temp mmap arrays is being deleted. All existing temp mmapp arrays will be unusable!
WARNING:root:New temp folder location. Temp mmap arrays are written to /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test_parallel/temp_mmap_urbkr3dy. Cleanup of this folder is OS dependant, and might need to be triggered manually!
Alignment complete.
assembling mosaic with shape (3, 3040, 3038)
created tempmmap array for assembled mosaic at /Users/sophia/Documents/GitHub/scPortrait/example_data/example_stitching/stitching_test_parallel/temp_mmap_urbkr3dy/temp_mmap_2664334953454325339.hdf
assembling channels with 3 workers
assembling mosaic: 0%| | 0/3 [00:00<?, ?it/s]
assembling channel 0: 100%|██████████| 9/9 [00:01<00:00, 7.81it/s]
assembling channel 2: 100%|██████████| 9/9 [00:01<00:00, 7.79it/s]
assembling channel 1: 100%|██████████| 9/9 [00:01<00:00, 7.78it/s]
assembling mosaic: 100%|██████████| 3/3 [00:01<00:00, 2.59it/s]
[17]:
#the stitched image is saved in the stitcher object and can be accessed via stitcher.assembled_mosaic
stitcher.assembled_mosaic
[17]:
|
||||||||||||||||
[18]:
# the stitched image can then be written to a variety of output formats
stitcher.write_tif(export_xml=True)
#alternatively the stitched images can also be written out to tifs in a multi-threaded format
#speedups here are limited by write speed to disk, the number of threads is limited by the number of channels available
stitcher.write_tif_parallel(export_xml=True)
stitcher.write_ome_zarr()
writing tif files: 100%|██████████| 3/3 [00:00<00:00, 212.63it/s]
Visualize Stitching Output
[19]:
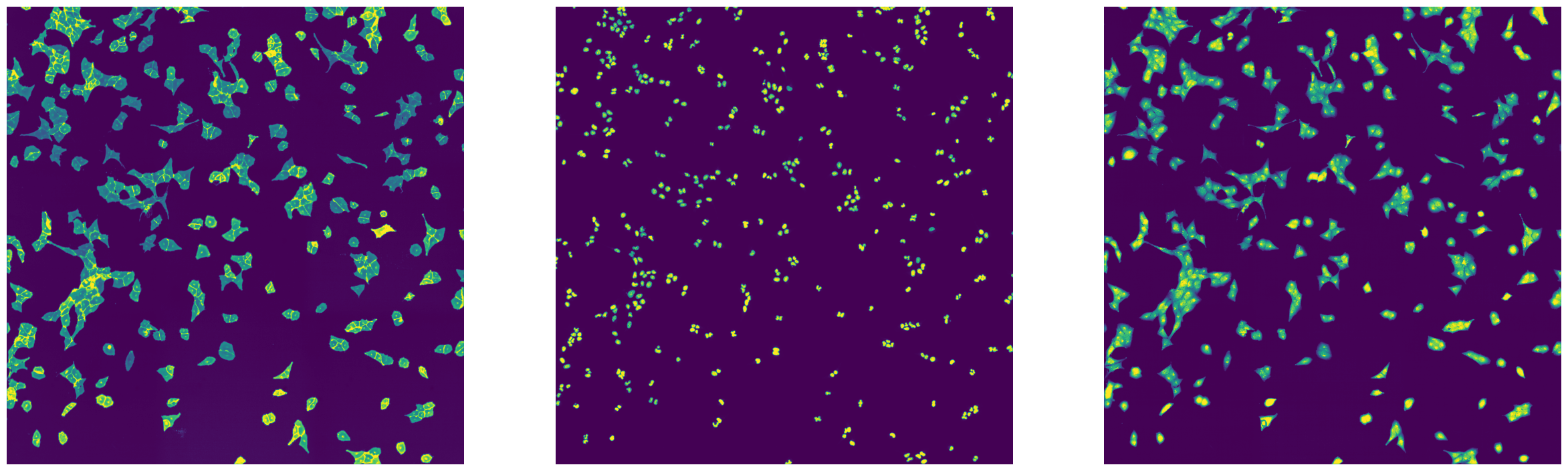
import matplotlib.pyplot as plt
from tifffile import imread
fig, axs = plt.subplots(1, 3, figsize = (30, 10))
axs[0].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_Alexa488.tif"))
axs[0].axis("off")
axs[1].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_DAPI.tif"))
axs[1].axis("off")
axs[2].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_mCherry.tif"))
axs[2].axis("off")
[19]:
(-0.5, 3037.5, 3039.5, -0.5)
[20]:
# compare parallel and non-parallel stitching
imread(f"{outdir_parallel}/stitching_test_parallel_Alexa488.tif") == imread(f"{outdir}/stitching_test_Alexa488.tif")
[20]:
array([[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]])
[21]:
del stitcher
[ ]:
[ ]: