scPortrait – image-based single cell analysis at scale in Python#
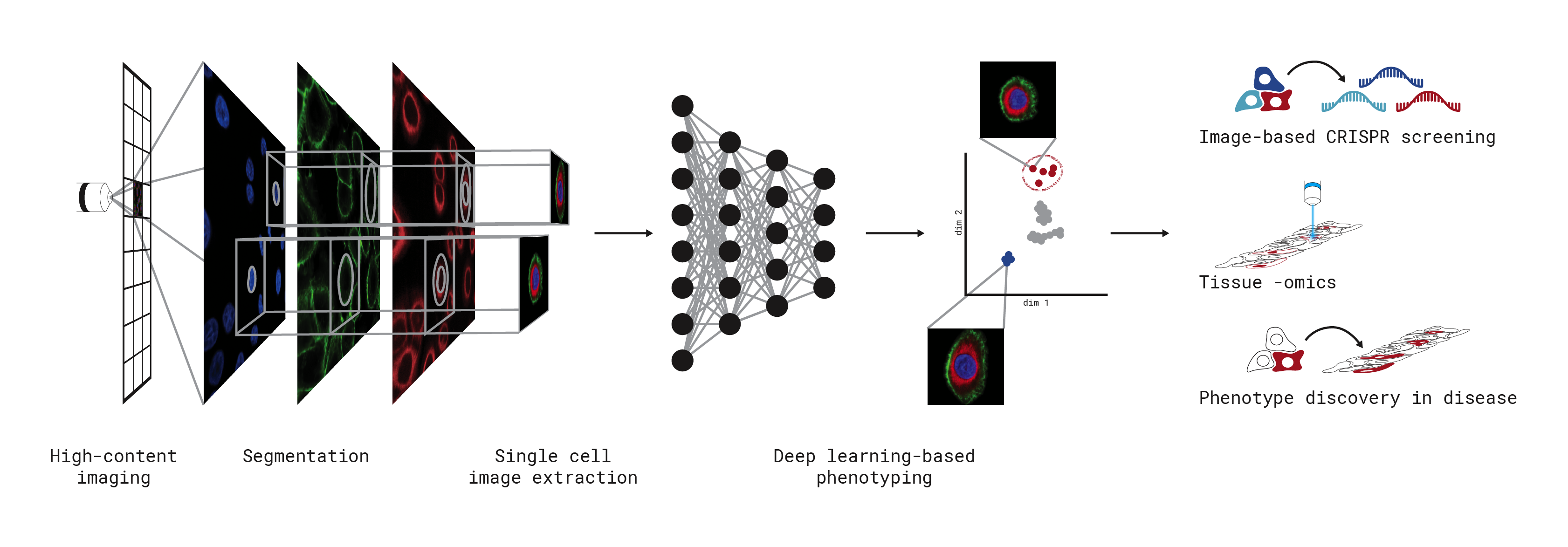
scPortrait is a scalable toolkit to analyse single-cell image datasets. This Python implementation efficiently segments individual cells, generates single-cell datasets and provides tools for the efficient deep learning classification of their phenotypes for downstream applications.
Installation#
Check out the installation instructions here. You can validate your installation by running one of the example notebooks here.
Getting Started#
You can check out our worfklow overview or our in-depth tutorial to get started with scPortrait. For more detailed information on the package, checkout the more detailed guides here. You can find a collection of example projects which run on datasets provided within scPortrait to help you get started with here. If you encounter issues feel free to open up a git issue.
Citing our Work#
scPortrait was developed by Sophia Mädler and Niklas Schmacke in the labs of Matthias Mann, Veit Hornung and Fabian Theis in 2024 and is actively being developed. If you are interested in contributing please reach out to the developers.
If you use our code please cite the following manuscript:
scPortrait integrates single-cell images into multimodal modeling. Sophia Clara Mädler, Niklas Arndt Schmacke, Alessandro Palma, Altana Namsaraeva, Ali Oğuz Can, Varvara Varlamova, Lukas Heumos, Mahima Arunkumar, Georg Wallmann, Veit Hornung, Fabian J. Theis, Matthias Mann bioRxiv 2025.09.22.677590; doi: https://doi.org/10.1101/2025.09.22.677590
Contributing#
We are excited for people to adapt and extend scPortrait to their needs. If you are interested in contributing to scPortrait, please reach out to the developers or open a pull request on our GitHub repository.
Documentation#
Ecosystem
Code Examples
Module API
- pipeline
- project
ProjectProject.configProject.nuc_seg_nameProject.cyto_seg_nameProject.sdata_pathProject.filehanderProject.DEFAULT_IMAGE_DTYPEProject.DEFAULT_SEGMENTATION_DTYPEProject.DEFAULT_SINGLE_CELL_IMAGE_DTYPEProject.sdataProject.update_featurization_f()Project.print_project_status()Project.view_sdata()Project.plot_input_image()Project.plot_he_image()Project.plot_segmentation_masks()Project.plot_single_cell_images()Project.load_input_from_array()Project.load_input_from_tif_files()Project.load_input_from_omezarr()Project.load_input_from_dask()Project.load_input_from_sdata()Project.complete_segmentation()Project.extract()Project.select()
- segmentation
- segmentation workflows
WGASegmentationShardedWGASegmentationDAPISegmentationShardedDAPISegmentationDAPISegmentationCellposeShardedDAPISegmentationCellposeCytosolSegmentationCellposeShardedCytosolSegmentationCellposeCytosolOnlySegmentationCellposeShardedCytosolOnlySegmentationCellposeCytosolSegmentationDownsamplingCellposeShardedCytosolSegmentationDownsamplingCellposeCytosolOnlySegmentationDownsamplingCellposeShardedCytosolOnlySegmentationDownsamplingCellpose
- extraction
- Featurization
- selection
- processing
- plotting
- io
- tools