Segmentation#
To ensure overall flexibility, scPortrait seperates code implementing the segmentation framework (i.e. how input data is loaded, segmentation methods are called or results saved) from the code implementing the actual segmentation algorithm (i.e. how the segmentation mask is calculated for a given input). This allows you to easily exchange one segmentation algorithm for another while retaining the rest of the code framework.
Segmentation frameworks are implemented as so-called segmentation classes and segmentation algorithms are implemented as so-called segmentation workflows. Each segmentation class is optimized for a given input data format and level of parallelization, and each workflow implements a different segmentation algorithm (e.g. thresholding based segmentation or deep learning based segmentation).
Segmentation classes#
scPortrait currently implements two different segmentation classes for each of the input data formats: a serialized segmentation class and a parallelized segmentation class. The serialized segmentation class is ideal for segmenting small input images in a single process. The parallelized segmentation classes can process larger-than-memory input images over multiple CPU cores.
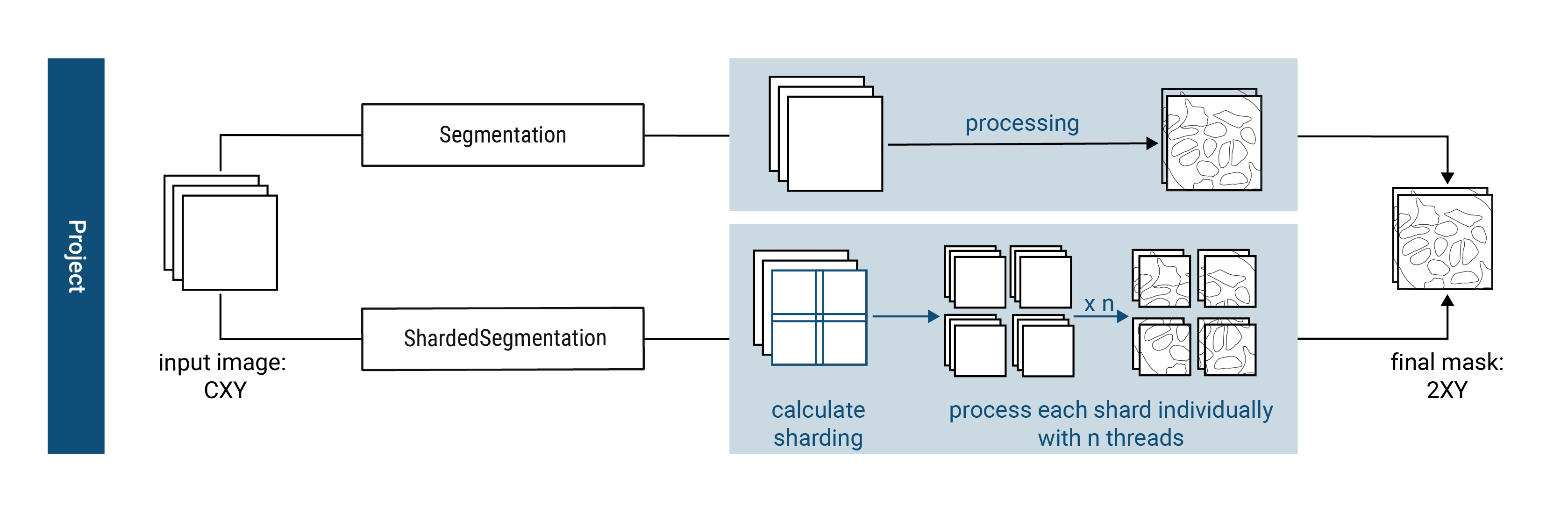
1. Segmentation#
The Segmentation class is optimized for processing input images of the format CXY within the context of a base scPortrait Project. It loads the input image into memory and then segments the image using the provided segmentation workflow. The resulting segmentation mask is then saved to disk.
Segmentation Workflows#
Within scPortrait a segmentation workflow refers to a specific segmentation algorithm that can be called by one of the segmentation classes described above. Currently the following segmentation workflows are available for each of the different segmentation classes. They are explained in more detail below:
If none of these segmentation approaches suit your particular needs you can easily implement your own workflow. In case you need help, please open a git issue.
Workflow overview#
Test goes here.
Configuring a segmentation workflow#
Workflow specific parameters are stored in config files#
The specific behaviour of a segmentation workflow is determined by the parameters in the supplied config file that is used to initialize the project. While different segmentation methods each have unique parameters that are required for the selected segmentation algorithm, all workflows share some common keys and a common structure.
Here is a strongly simplified config for a generic scPortrait Segmentation Workflow:
{SegmentationWorkflow}:
cache: "/path/to/directory/to/use/for/memorymapping/intermediate/results
nucleus_segmentation:
# parameters specific to nucleus segmentation method go here
key: value
# if `filter_masks_size` is set to True then the min and max size in px for each nucleus mask can be configured through these parameters
min_size: 200
max_size: 30000
cytosol_segmentation
# parameters specific to cytosol segmentation method go here
key: value
# if `filter_masks_size` is set to True then the min and max size in px for each cytosol mask can be configured through these parameters
min_size: 200
max_size: 30000
match_masks: True
filtering_threshold_mask_matching: 0.95
filter_masks_size: False
Methods that only perform a nucleus or cytosol segmentation step will only need to provide the relevant parameters for the step that is executed.
As for all scPortrait configs, they can contain a mix of mandatory as well as optional parameters. If an optional parameter is not specified within a given config, scPortrait will use the default value for it. For some parameters there are no default values implemented, so its absolutely mandatory that you provide these yourself. In case you try and execute a run with an incomplete config (i.e. where a mandatory parameter is missing) scPortrait will inform you of this so that you can update your config file accordingly.
Parameter |
Description |
Optional |
Default Value |
|---|---|---|---|
|
Specifies the directory to be used for out-of-memory backed computations. |
True |
Uses the current working directory, it is highly recommended to pass a specific directory though that is located on a fast-access drive (SSD). |
|
Contains all parameters specific to the nuclear segmentation step. |
||
|
Contains all parameters specific to the cytosolic segmentation step. |
||
|
Determines if the resulting masks should be filtered according to size, with min/max cutoffs specified per segmentation step. |
True |
False |
|
The minimum size in px that a mask needs to have to pass mask size filtering if |
Not optional if |
None |
|
The maximum size in px that a mask needs to have to pass mask size filtering if |
Not optional if |
None |
|
Specifies whether cytosolic and nuclear segmentation masks should be matched. If enabled, cytosol masks that do not match exactly one nuclear mask are removed, and vice versa. |
True |
True for methods that generate both a nuclear as well as a cytosol mask |
|
Defines the percentage of overlap required between a nuclear mask and a cytosol mask for them to be considered a match. |
True |
0.95 |
Input Channels Required for Segmentation Algorithm#
To generate segmentation masks, you will usually not require all of the channels present in your input images.
The different segmentation workflows will automatically subset the provided input images to only run on the channels of interest. This ensures efficient computation, as only the absolutely required information is loaded into memory while everything else is left on disk.
Depending on the segmentation method, either 1 or 2 input channels will be required to generate a segmentation mask.
Segmentation Method |
Number of Input Channels |
Number of Output Masks |
|---|---|---|
2 |
2 |
|
1 |
1 |
|
2 |
2 |
|
1 |
1 |
|
2 |
1 |
To automatically select the relevant input channels for segmentation, scPortrait assumes that you have loaded your input channels in the following order:
Nuclear marker channel
Cell membrane marker channel
All other channels
In some cases, you may want to customize this behavior, e.g., if you want to use a maximum-intensity projection of multiple input channels as a proxy for segmentation.
This behavior can also be modified through the configuration file. Below, we will illustrate a few different use cases.
Case 1: combine multiple channels through maximum-intensity projection#
By adding a key with either combine_nucleus_channels or combine_cytosol_channels which provides a list of channel indices to combine you can perform a maximum intensity projection of the provided channel indexes before passing the newly generated channel to the respective segmentation algorithm.
cache: "/path/to/directory/to/use/for/memorymapping/intermediate/results
nucleus_segmentation:
# parameters specific to nucleus segmentation method go here
key: value
cytosol_segmentation
# parameters specific to cytosol segmentation method go here
key: value
combine_nucleus_channels: [0, 2]
combine_cytosol_channels: [1, 2]
Case 2: select different channel ids for segmentation because your channel order differs to the expected format#
You can override the default behaviour by manually suppling specific channel index ids that contain nuclear or cytoplasmic information.
cache: "/path/to/directory/to/use/for/memorymapping/intermediate/results
nucleus_segmentation:
# parameters specific to nucleus segmentation method go here
key: value
cytosol_segmentation
# parameters specific to cytosol segmentation method go here
key: value
segmentation_channel_nuclei: [2]
segmentation_channel_cytosol: [2]
Case 3: do a combination of the two#
Both use cases can of course also be combined. In case you pass both combine_{mask_name}_channels and segmentation_channel_{mask_name} with differing values, combine_channel_{mask_name} will superscede segmentation_channel_{mask_name}.
cache: "/path/to/directory/to/use/for/memorymapping/intermediate/results
nucleus_segmentation:
# parameters specific to nucleus segmentation method go here
key: value
cytosol_segmentation
# parameters specific to cytosol segmentation method go here
key: value
segmentation_channel_nuclei: [2]
combine_cytosol_channels: [1, 2]
Customize Cellpose Model Behaviour#
You can customize the specific behaviour of all cellpose models via the method specific config file.
{mask_name}_segmentation:
model: "cyto2"
model_path: "path/to/a/custom/cellpose/model"
normalize: True
diameter: None
resample: True
rescale: None
flow_threshold: 0.4
cellprob_threshold: 0.0
The indicated keys are wrappers for the parameters of cellpose.models.CellposeModel.eval and have the same function.
Parameter |
Description |
Optional |
Default Value |
|---|---|---|---|
|
Name of a built-in Cellpose model. |
Only if |
|
|
Path to a custom trained Cellpose model. |
True |
|
|
Wrapper for Cellpose |
True |
|
|
Wrapper for Cellpose |
True |
|
|
Wrapper for Cellpose |
True |
|
|
Wrapper for Cellpose |
True |
|
|
Wrapper for Cellpose |
True |
|
|
Wrapper for Cellpose |
True |
|
Detailed Workflow Descriptions#
WGA segmentation#
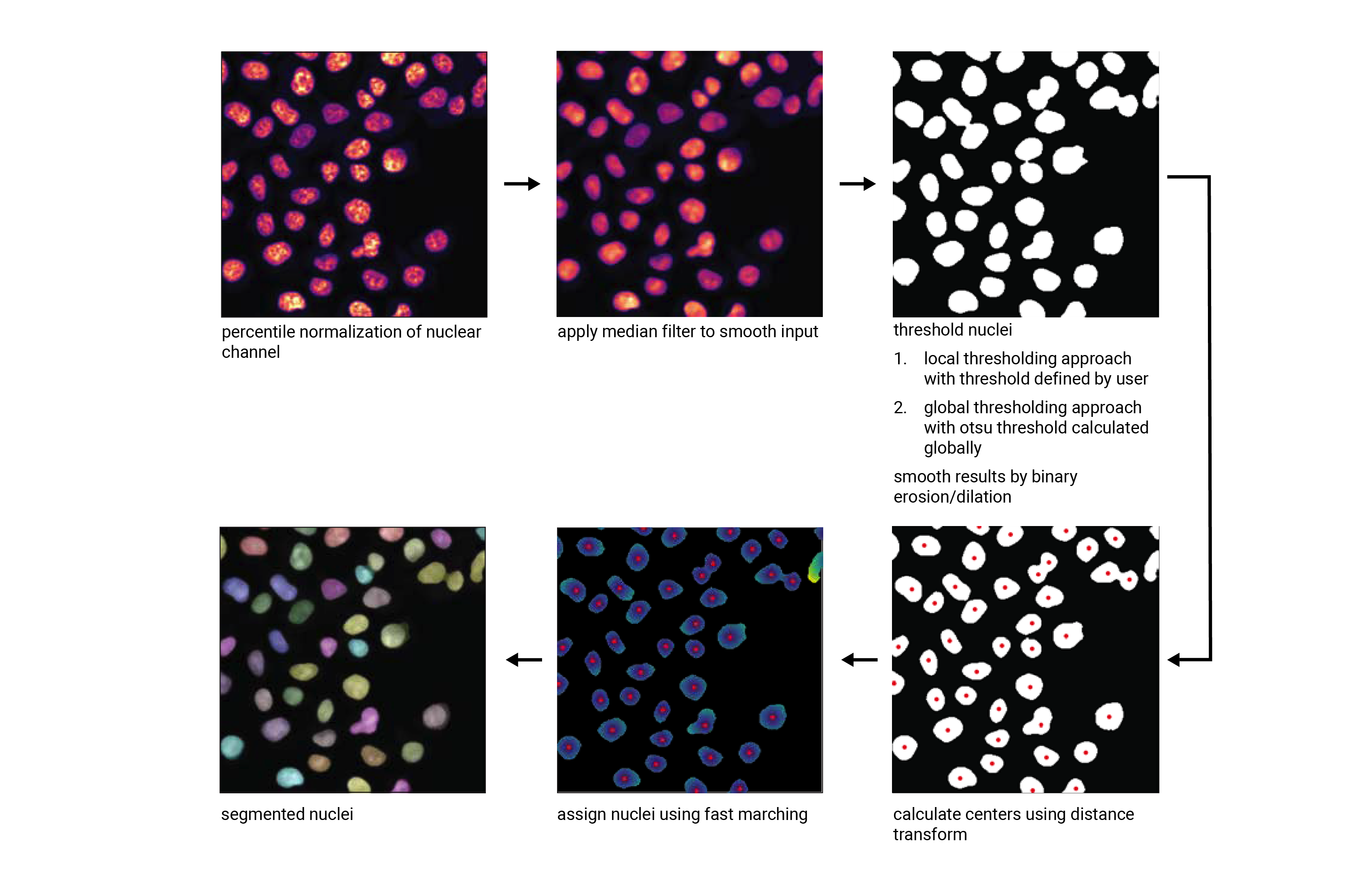
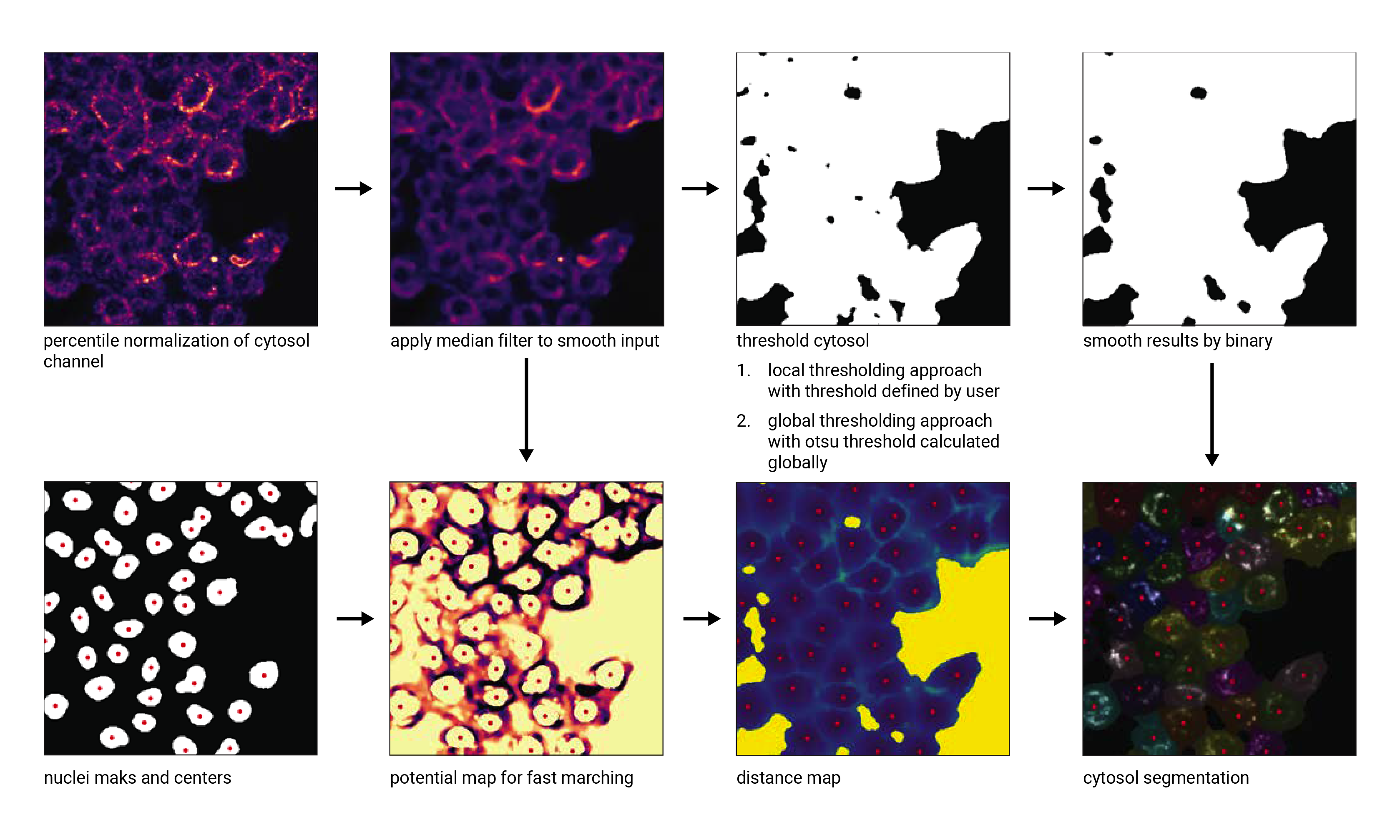
This segmentation workflow aims to segment mononucleated cells, i.e. cells that contain exactly one nucleus. Based on a nuclear stain and a cellmembrane stain, it first uses a thresholding approach to identify nuclei which are assumed to be the center of each cell. Then in a second step, the center of the identified nuclei are used as a starting point to generate a potential map using the cytosolic stain. This potential map is then used to segment the cytosol using a watershed approach. At the end of the workflow the user obtains both a nuclear and a cytosolic segmentation mask where each cytosol is matched to exactly one nucleus as kann be identified by the matching cell id.
This segmentation workflow is implemented to only run on the CPU. As such it can easily be scaled up to run on large datasets using parallel processing over multiple cores using either the ShardedSegmentation class or the MultithreadedSegmentation class respectively. However, it has a lot of parameters that need to be adjusted for different datasets to obtain an optimal segmentation.
WGASegmentation:
lower_quantile_normalization: 0.001
upper_quantile_normalization: 0.999
median_filter_size: 4 # Size in pixels
nucleus_segmentation:
lower_quantile_normalization: 0.01 # quantile normalization of dapi channel before local tresholding. Strong normalization (0.05,0.95) can help with nuclear speckles.
upper_quantile_normalization: 0.99 # quantile normalization of dapi channel before local tresholding. Strong normalization (0.05,0.95) can help with nuclear speckles.
median_block: 41 # Size of pixel disk used for median, should be uneven
median_step: 4
threshold: 0.2 # threshold above local median for nuclear segmentation
min_distance: 8 # minimum distance between two nucleis in pixel
peak_footprint: 7 #
speckle_kernel: 9 # Erosion followed by Dilation to remove speckels, size in pixels, should be uneven
dilation: 0 # final dilation of pixel mask
min_size: 200 # minimum nucleus area in pixel
max_size: 1000 # maximum nucleus area in pixel
contact_filter: 0.5 # minimum nucleus contact with background
cytosol_segmentation:
threshold: 0.05 # treshold above which cytosol is detected
lower_quantile_normalization: 0.01
upper_quantile_normalization: 0.99
erosion: 2 # erosion and dilation are used for speckle removal and shrinking / dilation
dilation: 7 # for no change in size choose erosion = dilation, for larger cells increase the mask erosion
min_clip: 0
max_clip: 0.2
min_size: 200
max_size: 6000
chunk_size: 50
filter_masks_size: True
Nucleus Segmentation Algorithm#
Cytosol Segmentation Algorithm#
DAPI segmentation#
This segmentation workflow aims to only segment nuclei. Based on a nuclear stain, it uses the same thresholding approach used during the WGA segmentation to identify nuclei. To ensure compatability with the downstream extraction workflow which assumes the presence of both a nuclear and a cytosolic segmentation mask the nuclear mask is duplicated and also used as the cytosolic mask. The generated single cell datasets using this segmentation method only focus on signals contained within the nuclear region.
DAPISegmentation:
input_channels: 3
chunk_size: 50 # chunk size for chunked HDF5 storage. is needed for correct caching and high performance reading. should be left at 50.
lower_quantile_normalization: 0.001
upper_quantile_normalization: 0.999
median_filter_size: 4 # Size in pixels
nucleus_segmentation:
lower_quantile_normalization: 0.01 # quantile normalization of dapi channel before local tresholding. Strong normalization (0.05,0.95) can help with nuclear speckles.
upper_quantile_normalization: 0.99 # quantile normalization of dapi channel before local tresholding. Strong normalization (0.05,0.95) can help with nuclear speckles.
median_block: 41 # Size of pixel disk used for median, should be uneven
median_step: 4
threshold: 0.2 # threshold above which nucleus is detected, if not specified a global threshold is calcualted using otsu
min_distance: 8 # minimum distance between two nucleis in pixel
peak_footprint: 7 #
speckle_kernel: 9 # Erosion followed by Dilation to remove speckels, size in pixels, should be uneven
dilation: 0 # final dilation of pixel mask
min_size: 200 # minimum nucleus area in pixel
max_size: 5000 # maximum nucleus area in pixel
contact_filter: 0.5 # minimum nucleus contact with background
chunk_size: 50
Nucleus Segmentation Algorithm#

Cytosol Cellpose segmentation#
This segmentation workflow is built around the cellular segmentation algorithm cellpose . Cellpose is a deep neural network with a U-net style architecture that was trained on large datasets of microscopy images of cells. It provides very accurate out of the box segmentation models for both nuclei and cytosols but also allows you to fine-tune models using your own data.
The scPortrait implementation of the cellpose segmenation algorithm allows you to perform both a nuclear and cytosolic segmentation and align the cellids between the two resulting masks. This means that the nucleus and the cytosol belonging to the same cell have the same cellids. Furthermore, it performs some filtering steps to remove the masks from multi-nucleated cells or those with only a nuclear or cytosolic mask. This ensures that only cells which show a normal physiology are retained for further analysis.
While this segmentation workflow is also capable of running on a CPU it is highly recommended to utilize a GPU for better performance. If your system has more than one GPU available, in a ShardedSegmentation context, you can specify the number of GPUs to be used via the configuration file (nGPUs).
If you utilize this segmentation workflow please also consider citing the cellpose paper.
ShardedCytosolSegmentationCellpose:
shard_size: 2000000 # maxmimum number of pixel per tile
overlap_px: 100
nGPUs: 1
threads: 2 # number of shards / tiles segmented at the same size. should be adapted to the maximum amount allowed by memory.
cache: "."
nucleus_segmentation:
model: "nuclei"
cytosol_segmentation:
model: "cyto2"
match_masks: True
filter_masks_size: False
DAPI Cellpose segmentation#
This segmentation workflow is also built around the cellular segmentation algorithm cellpose but only performs a nuclear segmentation. This algorithm only takes a single input channel to generate a single output mask. The generated single cell datasets using this segmentation method only focus on signals contained within the nuclear region.
As for the cytosol segmentation cellpose workflow it is highly recommended to utilize a GPU. If your system has more than one GPU available, in a ShardedSegmentation context, you can specify the number of GPUs to be used via the configuration file (nGPUs).
If you utilize this segmentation workflow please also consider citing the cellpose paper.
ShardedDAPISegmentationCellpose:
#segmentation class specific
input_channels: 2
output_masks: 2
shard_size: 120000000 # maxmimum number of pixel per tile
overlap_px: 100
chunk_size: 50 # chunk size for chunked HDF5 storage. is needed for correct caching and high performance reading. should be left at 50.
cache: "/fs/pool/pool-mann-maedler-shared/temp"
# segmentation workflow specific
nGPUs: 2
lower_quantile_normalization: 0.001
upper_quantile_normalization: 0.999
median_filter_size: 6 # Size in pixels
nucleus_segmentation:
model: "nuclei"
Cytosol Only Cellpose segmentation#
This segmentation workflow is also built around the cellular segmentation algorithm cellpose but only performs a cytosol segmentation. Unlike the DAPI segmentation cellpose workflow it uses two input channels to generate a single output mask. The generated single cell datasets using this segmentation method will contain all signal from within the cytosolic region.
As for the cytosol segmentation cellpose workflow it is highly recommended to utilize a GPU. If your system has more than one GPU available, in a ShardedSegmentation context, you can specify the number of GPUs to be used via the configuration file (nGPUs).
If you utilize this segmentation workflow please also consider citing the cellpose paper.
ShardedCytosolOnlySegmentationCellpose:
shard_size: 2000000 # maxmimum number of pixel per tile
overlap_px: 100
nGPUs: 1
threads: 2 # number of shards / tiles segmented at the same size. should be adapted to the maximum amount allowed by memory.
cache: "."
cytosol_segmentation:
model: "cyto2"
match_masks: True
filter_masks_size: False