Stitch individual tif files into whole-slide images#
Stitcing in scPortrait is built on top of the Ashlar package.
When stitching from .tif files, Ashlar reads channel and tile position information from filenames according to a predefined pattern. Hence, filenames matter when stitching from .tif files.
[1]:
import os
import numpy as np
import matplotlib.pyplot as plt
from scportrait.data._datasets import dataset_stitching_example
from scportrait.tools.stitch import ParallelStitcher, Stitcher
/Users/sophia/mambaforge/envs/scportrait/lib/python3.11/site-packages/xarray_schema/__init__.py:1: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
from pkg_resources import DistributionNotFound, get_distribution
Single-threaded Stitching#
Initializing the Stitcher object#
[2]:
input_dir = dataset_stitching_example()
slidename = "stitching_test"
outdir = os.path.join(str(input_dir).replace("stitching_example", "example_projects/stitching"), slidename)
row = str(2).zfill(2) # specify the row of the well you want to stitch, here = 2
well = str(4).zfill(2) # specifc the well number you wish to stitch, here = 4
zstack_value = str(1).zfill(
3
) # specify the zstack you want to stitch. for multiple zstacks please make a loop and iterate through each of them.
timepoint = str(1).zfill(3) # specify the timepoint you wish to stitch
pattern = f"Timepoint{timepoint}_Row{row}_Well{well}_{{channel}}_zstack{zstack_value}_r{{row:03}}_c{{col:03}}.tif"
# initialize stitcher
stitcher = Stitcher(
os.path.abspath(input_dir),
slidename,
outdir,
pattern=pattern,
stitching_channel="Alexa488",
channel_order=[
"DAPI",
"Alexa488",
"mCherry",
], # this can be used to override the order of channels in the final stitched image for image types like ome.zarr, if not specified alphabetical order is used
overlap=0.1,
max_shift=30,
filter_sigma=0,
rescale_range={"Alexa488": (1, 99), "DAPI": (1, 99), "mCherry": (1, 99)},
overwrite=True,
image_dtype=np.uint16,
)
Downloading...: 100%|██████████| 42.0M/42.0M [00:37<00:00, 1.11MB/s]
Output directory created at: /Users/sophia/Documents/GitHub/scPortrait/src/scportrait/scportrait_data/example_projects/stitching/stitching_test
You can access information on which channels are present in your dataset and the specific values of the parameters relevant to stitching by executing:
[3]:
stitcher.get_stitching_information()
Tile positions will be calculated based on channel: Alexa488
Channel Names: ['Alexa488', 'DAPI', 'mCherry']
Overlap of image tiles: 0.1
Max Shift value: 30
Filter Sigma value: 0
Output will be written to: /Users/sophia/Documents/GitHub/scPortrait/src/scportrait/scportrait_data/example_projects/stitching/stitching_test
Generating thumbnails#
[4]:
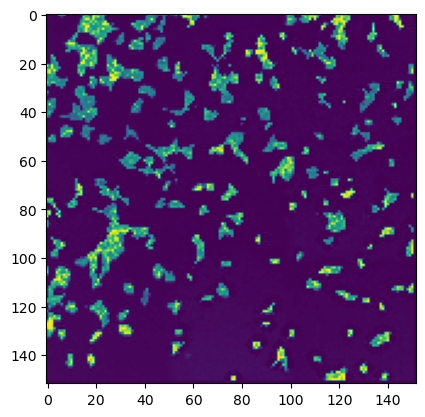
stitcher.generate_thumbnail()
assembling thumbnail 9/9
[5]:
# thumbnail is saved in the stitcher object and can be accessed via stitcher.thumbnail
plt.imshow(stitcher.thumbnail)
[5]:
<matplotlib.image.AxesImage at 0x37f360e10>
[6]:
# alterantively it can be saved to a tif file
stitcher.write_thumbnail()
Generating full-scale stitched image#
[7]:
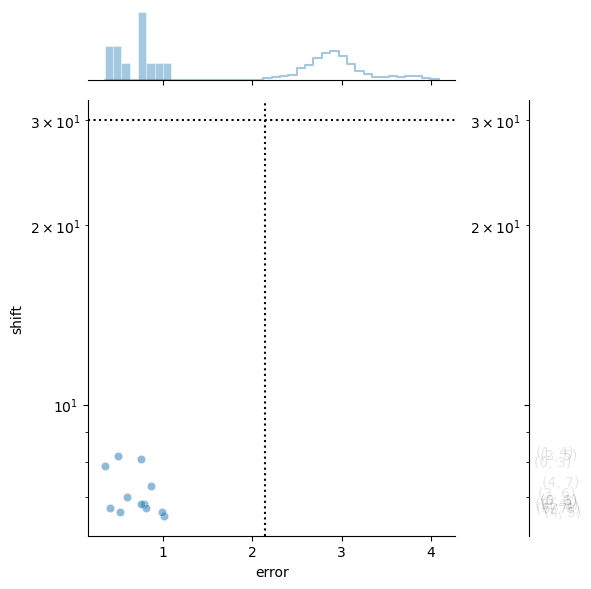
stitcher.stitch()
performing stitching on channel Alexa488 with id number 0
quantifying alignment error 1000/1000
aligning edge 12/12
Alignment complete.
current channel order: [0, 1, 2]
new channel order [1, 0, 2]
assembling mosaic with shape (3, 3040, 3038)
created tempmmap array for assembled mosaic at /Users/sophia/Documents/GitHub/scPortrait/src/scportrait/scportrait_data/example_projects/stitching/stitching_test/temp_mmap_32dgb04r/temp_mmap_2604633988772087563.hdf
merging tile 9/9
merging tile 9/9
merging tile 9/9
[8]:
# the stitched image is saved in the stitcher object and can be accessed via stitcher.assembled_mosaic
stitcher.assembled_mosaic
[8]:
|
||||||||||||||||
[9]:
# the stitched image can then be written to a variety of output formats
stitcher.write_tif(export_xml=True)
stitcher.write_ome_zarr()
[10]:
import matplotlib.pyplot as plt
from tifffile import imread
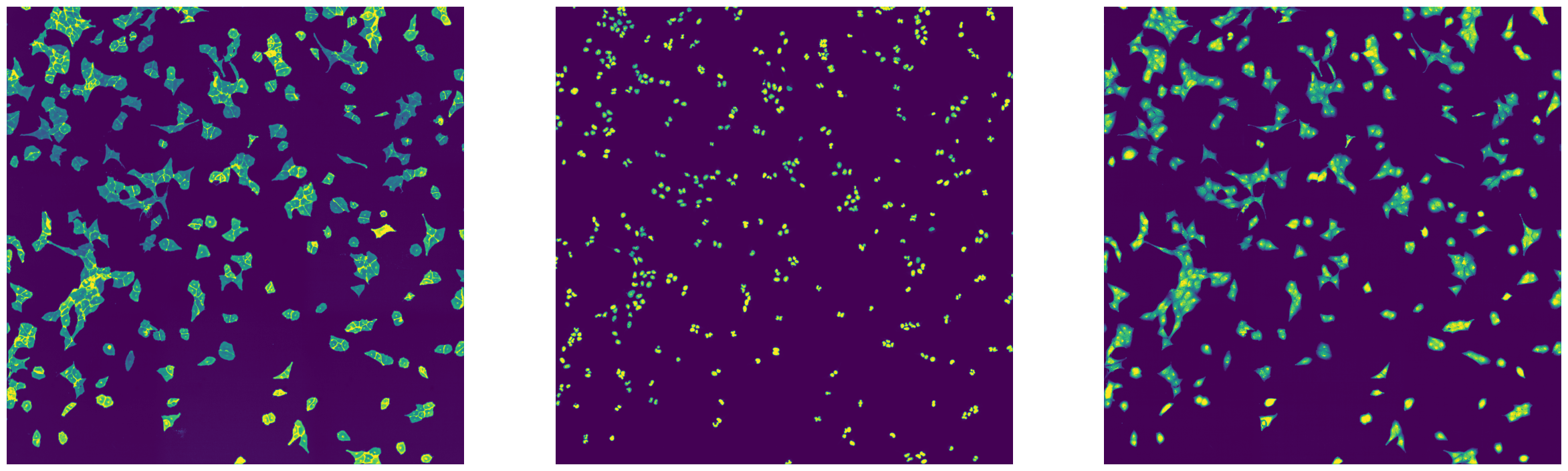
fig, axs = plt.subplots(1, 3, figsize=(30, 10))
axs[0].imshow(imread(f"{outdir}/stitching_test_Alexa488.tif"))
axs[0].axis("off")
axs[1].imshow(imread(f"{outdir}/stitching_test_DAPI.tif"))
axs[1].axis("off")
axs[2].imshow(imread(f"{outdir}/stitching_test_mCherry.tif"))
axs[2].axis("off")
[10]:
(-0.5, 3037.5, 3039.5, -0.5)
[11]:
del stitcher
Multi-threaded Stitching#
The ParallelStitcher class can speed up stitching by using multiple threads. The code to start stitching remains the same, but ParallelStitcher takes an additional argument threads, specifying the number of parallel threads to use. This multi-threaded implementation uses some of ashlars base functionality but implements custom functions for several processing steps.
Initializing the ParallelStitcher object#
[12]:
input_dir = dataset_stitching_example()
slidename = "stitching_test_parallel"
outdir_parallel = os.path.join(str(input_dir).replace("stitching_example", "example_projects/stitching"), slidename)
row = str(2).zfill(2) # specify the row of the well you want to stitch, here = 2
well = str(4).zfill(2) # specifc the well number you wish to stitch, here = 4
zstack_value = str(1).zfill(
3
) # specify the zstack you want to stitch. for multiple zstacks please make a loop and iterate through each of them.
timepoint = str(1).zfill(3) # specify the timepoint you wish to stitch
pattern = f"Timepoint{timepoint}_Row{row}_Well{well}_{{channel}}_zstack{zstack_value}_r{{row:03}}_c{{col:03}}.tif"
# initialize stitcher
stitcher = ParallelStitcher(
input_dir,
slidename,
outdir_parallel,
pattern=pattern,
stitching_channel="Alexa488",
channel_order=["DAPI", "Alexa488", "mCherry"],
overlap=0.1,
max_shift=30,
filter_sigma=0,
rescale_range={"Alexa488": (1, 99), "DAPI": (1, 99), "mCherry": (1, 99)},
overwrite=True,
threads=12,
)
Output directory created at: /Users/sophia/Documents/GitHub/scPortrait/src/scportrait/scportrait_data/example_projects/stitching/stitching_test_parallel
Generating thumbnails#
[13]:
stitcher.generate_thumbnail()
assembling thumbnail 1/9Setting dtype to uint16
assembling thumbnail 9/9
[14]:
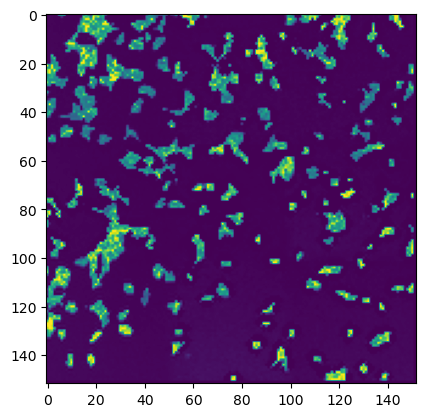
# thumbnail is saved in the stitcher object and can be accessed via stitcher.thumbnail
plt.imshow(stitcher.thumbnail)
[14]:
<matplotlib.image.AxesImage at 0x39ff39250>
[15]:
# alterantively it can be saved to a tif file
stitcher.write_thumbnail()
Generating full-scale stitched image#
[16]:
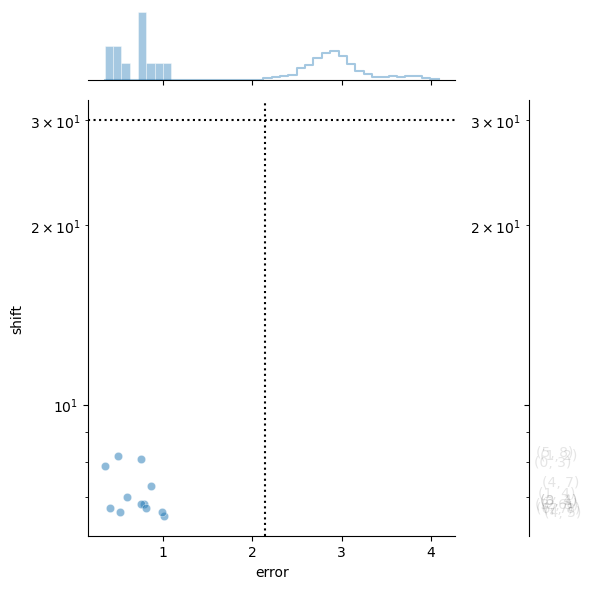
stitcher.stitch()
performing stitching on channel Alexa488 with id number 0
Setting dtype to uint16
using graph-tool to build spanning tree
Alignment complete.
current channel order: [0, 1, 2]
new channel order [1, 0, 2]
assembling mosaic with shape (3, 3040, 3038)
created tempmmap array for assembled mosaic at /Users/sophia/Documents/GitHub/scPortrait/src/scportrait/scportrait_data/example_projects/stitching/stitching_test_parallel/temp_mmap_qi1ziivg/temp_mmap_2008065058678754820.hdf
assembling channels with 3 workers
[17]:
# the stitched image is saved in the stitcher object and can be accessed via stitcher.assembled_mosaic
stitcher.assembled_mosaic
[17]:
|
||||||||||||||||
[18]:
# the stitched image can then be written to a variety of output formats
stitcher.write_tif(export_xml=True)
# alternatively the stitched images can also be written out to tifs in a multi-threaded format
# speedups here are limited by write speed to disk, the number of threads is limited by the number of channels available
stitcher.write_tif_parallel(export_xml=True)
stitcher.write_ome_zarr()
Visualize Stitching Output#
[19]:
import matplotlib.pyplot as plt
from tifffile import imread
fig, axs = plt.subplots(1, 3, figsize=(30, 10))
axs[0].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_Alexa488.tif"))
axs[0].axis("off")
axs[1].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_DAPI.tif"))
axs[1].axis("off")
axs[2].imshow(imread(f"{outdir_parallel}/stitching_test_parallel_mCherry.tif"))
axs[2].axis("off")
[19]:
(-0.5, 3037.5, 3039.5, -0.5)
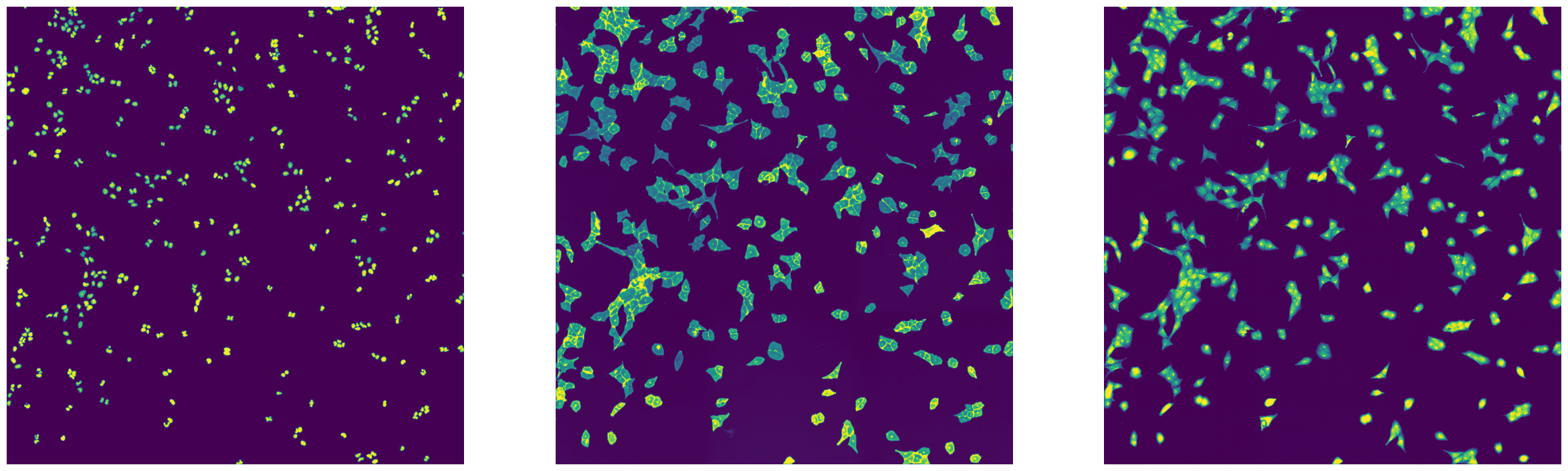
The results generated from a parallelized vs a single-threaded run are identical.
[20]:
# compare parallel and non-parallel stitching
imread(f"{outdir_parallel}/stitching_test_parallel_Alexa488.tif") == imread(f"{outdir}/stitching_test_Alexa488.tif")
[20]:
array([[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]])
[21]:
del stitcher
[ ]: